
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
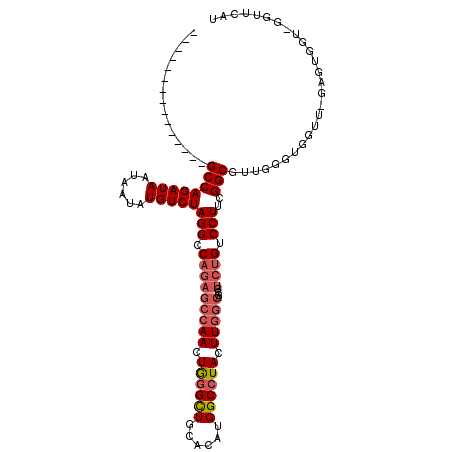
| Location | 18,326,793 – 18,326,920 |
| Length | 127 |
| Max. P | 0.685096 |

| Location | 18,326,793 – 18,326,889 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -24.72 |
| Energy contribution | -25.80 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
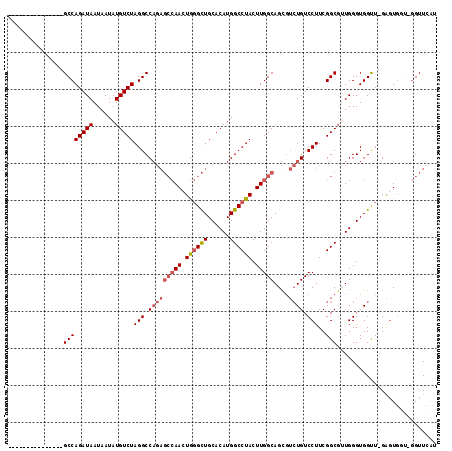
>3R_DroMel_CAF1 18326793 96 - 27905053 ---------------GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGUGUCUGUCCUUCGGCGUUGGGUGGUU-GAGUG----GUUCAU ---------------((((((((......))))).)))..((((((.((.(((..(.((..(((.....(((((...)))))....)))..)).)..)))-.))))----)))).. ( -35.00) >DroSec_CAF1 12335 100 - 1 ---------------GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGCGUCUGUCCUUCGGCGUUGGGUGGUU-GAGUGGUUGGUUCAU ---------------((((((((......))))).)))..((((((((((((((......)))))(((..((((((((........)))))).....)).-.)))))))))))).. ( -38.90) >DroSim_CAF1 12600 100 - 1 ---------------GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGCGUCUGUCCUUCGGCGUUGGGUGGUU-GAGUGGUGGGUUCAU ---------------((((((((......))))).)))..(((((.(((.(((..(.....(((.....)))((((((........))))))..)..)))-.)))....))))).. ( -36.30) >DroYak_CAF1 13107 94 - 1 -----------CC--GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAAAUGGGCUGCACAUGGCCUACUUGGCAGCGUCUGUCCUUCGGCGGUGGGUGGUU-GA-U----GGUU--- -----------((--((((((((......)))))(((.(((((((((.((((((......)))))).)))))....)))).)))..))))).........-..-.----....--- ( -35.20) >DroAna_CAF1 39313 110 - 1 AUGGUUGGAUGGUUGGCCAGAUAAUAAUAUGUCUAGGCC------AAAUACGUUGCACAUGGCCUACUUGGCAGUCGAUGUCCUUCGGCUGUGGGUGGCUCGGGUGGUCGGUUGGU .(..(((((((.(((((((((((......))))).))))------))...)))).(((..((((.((.(.((((((((......)))))))).)))))))...)))..)))..).. ( -40.20) >consensus _______________GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGCGUCUGUCCUUCGGCGUUGGGUGGUU_GAGUGGU_GGUUCAU ...............((((((((......)))))(((.(((((((((.((((((......)))))).)))))....)))).)))..)))........................... (-24.72 = -25.80 + 1.08)

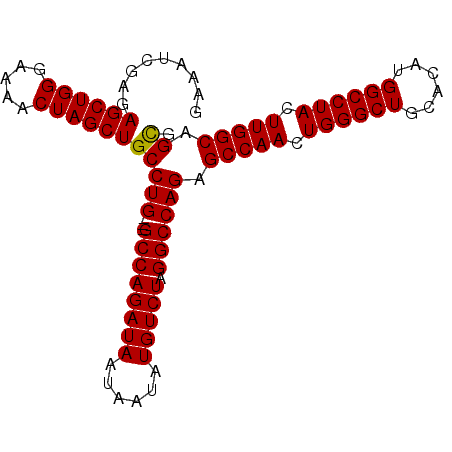
| Location | 18,326,828 – 18,326,920 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -35.54 |
| Energy contribution | -35.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
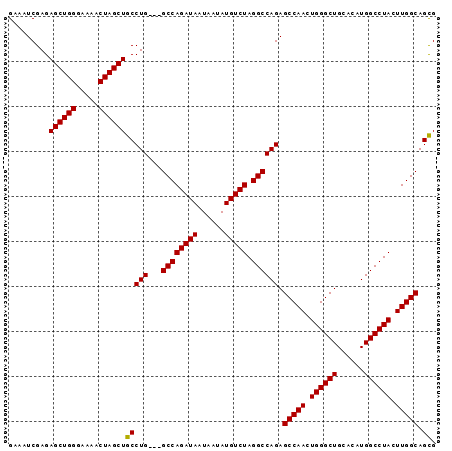
>3R_DroMel_CAF1 18326828 92 - 27905053 GGAAUCGAGAGCUGGGAAAACUAGCUGCCUG---GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGUG .........((((((.....))))))..(((---((((((((......))))).)))))).(((((.((((((......)))))).))))).... ( -40.90) >DroSec_CAF1 12374 92 - 1 GAAUUCGAGAGCUGGGAAAACUAGCUGCCUG---GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGCG .........((((((.....))))))(((((---((((((((......))))).)))))).(((((.((((((......)))))).))))).)). ( -42.40) >DroSim_CAF1 12639 92 - 1 GGAAUCGAGAGCUGGGAAAACUAGCUGCCUG---GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGCG .........((((((.....))))))(((((---((((((((......))))).)))))).(((((.((((((......)))))).))))).)). ( -42.40) >DroYak_CAF1 13138 95 - 1 GAAAUCGAGAGCUGGGAAAACUAGCUGCCUGGCCGCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAAAUGGGCUGCACAUGGCCUACUUGGCAGCG .........((((((.....))))))((((((((...(((((......))))).)))))).(((((.((((((......)))))).))))).)). ( -41.80) >consensus GAAAUCGAGAGCUGGGAAAACUAGCUGCCUG___GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUGGCAGCG .........((((((.....))))))(((((...((((((((......))))).)))))).(((((.((((((......)))))).))))).)). (-35.54 = -35.35 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:04 2006