| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,325,335 – 18,325,444 |
| Length | 109 |
| Max. P | 0.800800 |

| Location | 18,325,335 – 18,325,444 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
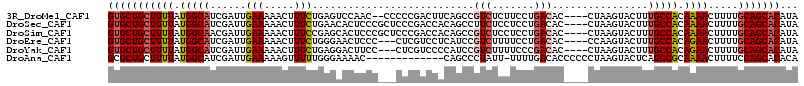
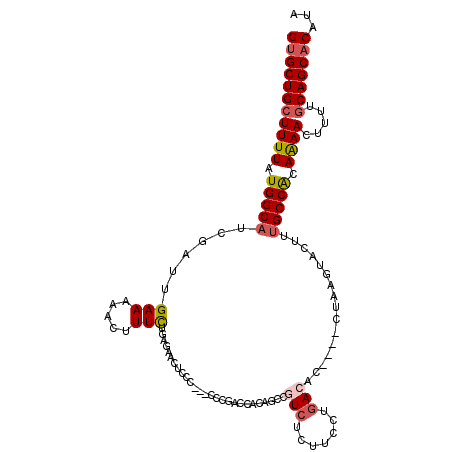
>3R_DroMel_CAF1 18325335 109 + 27905053 UAUGUGCUGCAAAAGUUUUGUGGCAAAGUACUUAG----GUGUCAGGAAGAGACGGCUGAAGUCGGGGG--GUUGGACUCAGAAAGUUUUUCAAUCGAUGCCAUAAAAGCAGCAC ...(((((((.....(((((((((.......((((----.((((.......)))).)))).(((((..(--(..(((((.....))))).))..))))))))))))))))))))) ( -37.10) >DroSec_CAF1 10921 111 + 1 UAUGUGCUGCAAAAGUUUUGUGGCAAAGUACUUAG----GUGUCAGGAGGAGAAGGCUGUGGUCGGGAGCGGGAGUGUUCAGAAAGUUUUUCAAUCGAUGCCAUAAAAGCAGCAC ...(((((((.....((((((((((..((((....----))).)..((...(((((((....((..(((((....))))).)).)))))))...))..))))))))))))))))) ( -33.60) >DroSim_CAF1 11164 111 + 1 UAUGUGCUGCAAAAGUUUUGUGGCAAAGUACUUAG----GUGUCAGGAGGAGACGGCUGUGGUCGGGAGCGGGAGUGCUCGGAAAGUUUUUCAAUCGUUGCCAUAAAAGCAGCAC ...(((((((.....((((((((((((((((((.(----.(.((....(....)(((....))))).).)..))))))).((((....)))).....)))))))))))))))))) ( -38.10) >DroEre_CAF1 11214 108 + 1 UAUGUGCUGCAAAAGUUCUGUGGCAAAGUACUUGG----GUGUCAGGAAAAGACGGAUGAGGACGAG---GGGAGUUCCCAGAAAGUUUUUCAAUCGAUGCCAUAAAAGCAGCAC ...(((((((.....((.(((((((.....((((.----....))))......(((.(((((((...---(((....))).....).)))))).))).))))))).))))))))) ( -32.40) >DroYak_CAF1 11603 108 + 1 UAUGUGCUGCAAAAGUUCUGUGGCAAAGUACUUAG----GUGUCGGGAAAAGACGGAUGGGGACGAG---GGAAGUCCUCAGAAAGUUUUUCAAUCGAUGCCAUAAAAGCAGCAC ...(((((((..((((.((.......)).)))).(----(((((((..((((((...(((((((...---....)))))))....))))))...))))))))......))))))) ( -41.90) >DroAna_CAF1 37862 101 + 1 UGUGUGCUGGAAAAGUUUUGCGGCUGAGUACUUAGGGGGGUGUCAAAA-AAUAGGGCUG-------------GUUUUCCCAAAAACUUUUUCAAUCGAUGCCAUAAAAGCAGCGC ...((((((((((((((((....((((....))))(((((..(((...-........))-------------)..))))).)))))))))))).....(((.......))))))) ( -24.40) >consensus UAUGUGCUGCAAAAGUUUUGUGGCAAAGUACUUAG____GUGUCAGGAAAAGACGGCUGAGGUCGGG___GGGAGUGCUCAGAAAGUUUUUCAAUCGAUGCCAUAAAAGCAGCAC ...(((((((.....((((((((((..((((........))).)..((((((.....((....................)).....))))))......))))))))))))))))) (-18.49 = -19.13 + 0.64)



| Location | 18,325,335 – 18,325,444 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18325335 109 - 27905053 GUGCUGCUUUUAUGGCAUCGAUUGAAAAACUUUCUGAGUCCAAC--CCCCCGACUUCAGCCGUCUCUUCCUGACAC----CUAAGUACUUUGCCACAAAACUUUUGCAGCACAUA (((((((((((.(((((..(.............(((((((....--.....))).))))..(((.......)))..----.......)..))))).)))).....)))))))... ( -27.60) >DroSec_CAF1 10921 111 - 1 GUGCUGCUUUUAUGGCAUCGAUUGAAAAACUUUCUGAACACUCCCGCUCCCGACCACAGCCUUCUCCUCCUGACAC----CUAAGUACUUUGCCACAAAACUUUUGCAGCACAUA (((((((((((.(((((..(...(((.....)))...........(((.........)))................----.......)..))))).)))).....)))))))... ( -19.90) >DroSim_CAF1 11164 111 - 1 GUGCUGCUUUUAUGGCAACGAUUGAAAAACUUUCCGAGCACUCCCGCUCCCGACCACAGCCGUCUCCUCCUGACAC----CUAAGUACUUUGCCACAAAACUUUUGCAGCACAUA (((((((((((.((((((.(...............((((......))))............(((.......)))..----.......).)))))).)))).....)))))))... ( -29.00) >DroEre_CAF1 11214 108 - 1 GUGCUGCUUUUAUGGCAUCGAUUGAAAAACUUUCUGGGAACUCCC---CUCGUCCUCAUCCGUCUUUUCCUGACAC----CCAAGUACUUUGCCACAGAACUUUUGCAGCACAUA (((((((((((.(((((....(((...........(((....)))---.............(((.......)))..----.)))......))))).)))).....)))))))... ( -27.80) >DroYak_CAF1 11603 108 - 1 GUGCUGCUUUUAUGGCAUCGAUUGAAAAACUUUCUGAGGACUUCC---CUCGUCCCCAUCCGUCUUUUCCCGACAC----CUAAGUACUUUGCCACAGAACUUUUGCAGCACAUA (((((((((((.(((((..(...............((((.....)---)))..........(((.......)))..----.......)..))))).)))).....)))))))... ( -28.00) >DroAna_CAF1 37862 101 - 1 GCGCUGCUUUUAUGGCAUCGAUUGAAAAAGUUUUUGGGAAAAC-------------CAGCCCUAUU-UUUUGACACCCCCCUAAGUACUCAGCCGCAAAACUUUUCCAGCACACA ((((((..((((.((......(..(((((....((((.....)-------------))).....))-)))..).....)).))))....))).)))................... ( -15.60) >consensus GUGCUGCUUUUAUGGCAUCGAUUGAAAAACUUUCUGAGAACUCCC___CCCGACCACAGCCGUCUCUUCCUGACAC____CUAAGUACUUUGCCACAAAACUUUUGCAGCACAUA (((((((((((.(((((......(((.....)))...........................(((.......)))................))))).)))).....)))))))... (-16.72 = -17.22 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:00 2006