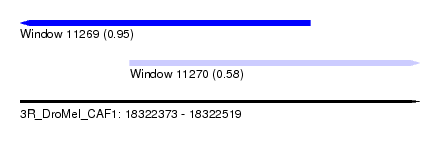
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,322,373 – 18,322,519 |
| Length | 146 |
| Max. P | 0.945601 |

| Location | 18,322,373 – 18,322,479 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.55 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -29.87 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
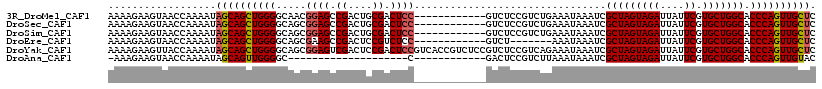
>3R_DroMel_CAF1 18322373 106 - 27905053 AAAAGAAGUAACCAAAAUAGCAGCUGGGGCAACGGAGCCGACUGCGACUCC------------GUCUCCGUCUGAAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGCUC ..................(((((((((((..((((((.((....)).))))------------))..))..............(((((((((....)).)))))))..))))))))). ( -38.90) >DroSec_CAF1 7993 106 - 1 AAAAGAAGUAACCAAAAUAGCAGCUGGGGCAGCGGAGCCGACUGCGACUCC------------GUCUCCGUCUGAAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGCUC ..................((((((((((.((((((((.((..........)------------).))))).))).........(((((((((....)).))))))).)))))))))). ( -41.10) >DroSim_CAF1 8184 106 - 1 AAAAGAAGUAACCAAAAUAGCAGCUGGGGCAGCGGAGCCGACUGCGACUCC------------GUCUCCGUCUGAAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGCUC ..................((((((((((.((((((((.((..........)------------).))))).))).........(((((((((....)).))))))).)))))))))). ( -41.10) >DroEre_CAF1 8320 99 - 1 AAAAGAAGUAACCAAAAUAGCAGCUGGGGCAGCGAAGCCGACUCCGUCUCC------------GUCU-------AAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGCUC ..................((((((((((...(((.((.((....)).)).)------------))..-------.........(((((((((....)).))))))).)))))))))). ( -32.50) >DroYak_CAF1 8659 118 - 1 AAAAGAAGUUACCAAAAUAGCAGCUGGGGCAGCGGAGUCGACUCCGACUCCGUCACCGUCUCCGUCUCCGUCAGAAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGCUC ..................(((((((((((..(((((((((....)))))))))..))..........................(((((((((....)).)))))))..))))))))). ( -44.50) >DroAna_CAF1 35045 85 - 1 -AAAGAAGUAACCAAAAUAGCAGUUGGGGC--------------------C------------GACUCCGUCUUAAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGUAC -..................(((..((((..--------------------.------------(((...)))...........(((((((((....)).))))))).))))..))).. ( -21.80) >consensus AAAAGAAGUAACCAAAAUAGCAGCUGGGGCAGCGGAGCCGACUGCGACUCC____________GUCUCCGUCUGAAAUAAAUCGCUAGUAGAUUAUUCGUGCUGGCACCCAGUUGCUC ..................((((((((((.....((((.((....)).))))................................(((((((((....)).))))))).)))))))))). (-29.87 = -30.30 + 0.43)
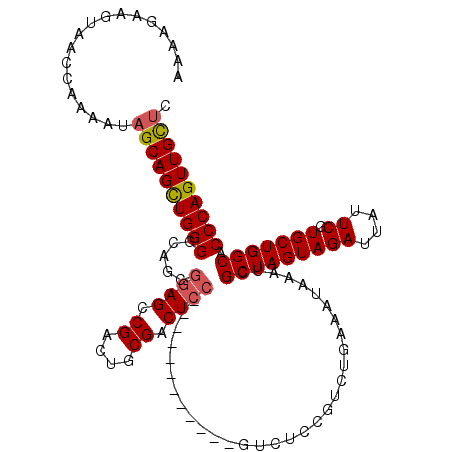
| Location | 18,322,413 – 18,322,519 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.34 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18322413 106 + 27905053 AUUUCAGACGGAGAC------------GGAGUCGCAGUCGGCUCCGUUGCCCCAGCUGCUAUUUUGGUUACUUCUUUUAUACACAAAUACCCAAAGAUUUGGGGGCAUGUUGGGUUCU ......(((((..((------------(((((((....)))))))))..))(((((((((..((((((............)).))))..(((((....))))))))).)))))))).. ( -37.00) >DroSec_CAF1 8033 106 + 1 AUUUCAGACGGAGAC------------GGAGUCGCAGUCGGCUCCGCUGCCCCAGCUGCUAUUUUGGUUACUUCUUUUAUACACAAAUACCCACAGAUUUGGGGGCAUGUUGGGUUCU .........(.((.(------------(((((((....)))))))))).)((((((((((..((((((............)).))))..((((......)))))))).)))))).... ( -35.20) >DroSim_CAF1 8224 105 + 1 AUUUCAGACGGAGAC------------GGAGUCGCAGUCGGCUCCGCUGCCCCAGCUGCUAUUUUGGUUACUUCUUUUAUACACAAAUACCCACAGAUUUGG-GGCAUGUUGGGUUCU .........(.((.(------------(((((((....)))))))))).)(((((((((...((((((............)).))))..((((......)))-)))).)))))).... ( -33.70) >DroEre_CAF1 8360 98 + 1 AUUU-------AGAC------------GGAGACGGAGUCGGCUUCGCUGCCCCAGCUGCUAUUUUGGUUACUUCUUUUAUACACAAAUACCCACGGAAUU-GGGGCAUGUUGGGUUCU ...(-------((.(------------((((.((....)).)))))))).(((((((((...((((((............)).))))..((((......)-)))))).)))))).... ( -26.20) >DroYak_CAF1 8699 118 + 1 AUUUCUGACGGAGACGGAGACGGUGACGGAGUCGGAGUCGACUCCGCUGCCCCAGCUGCUAUUUUGGUAACUUCUUUUAUACACAAAUACCCACAGCAUGAGGGGCAUGUUGGGUUUU ........((....))((((((((..((((((((....))))))))..)))((((((((...(((((((..........))).))))..(((.(.....).)))))).)))))))))) ( -41.50) >consensus AUUUCAGACGGAGAC____________GGAGUCGCAGUCGGCUCCGCUGCCCCAGCUGCUAUUUUGGUUACUUCUUUUAUACACAAAUACCCACAGAUUUGGGGGCAUGUUGGGUUCU ...........................(((((((....))))))).....((((((((((..((((((............)).))))..((((......)))))))).)))))).... (-25.62 = -26.34 + 0.72)

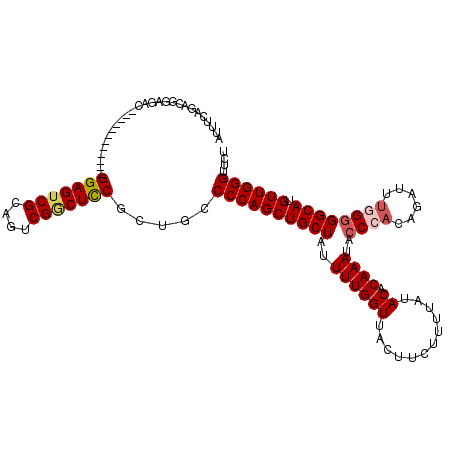
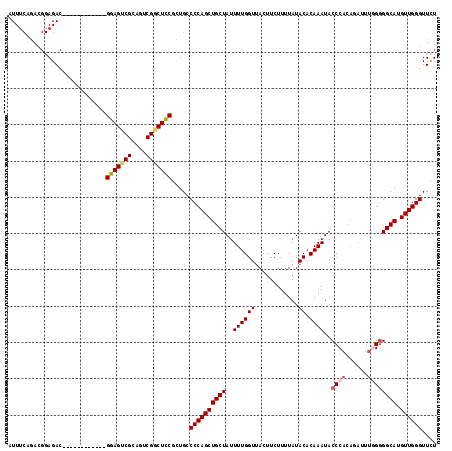
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:57 2006