
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,472,574 – 2,472,693 |
| Length | 119 |
| Max. P | 0.818678 |

| Location | 2,472,574 – 2,472,693 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -23.34 |
| Energy contribution | -25.38 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
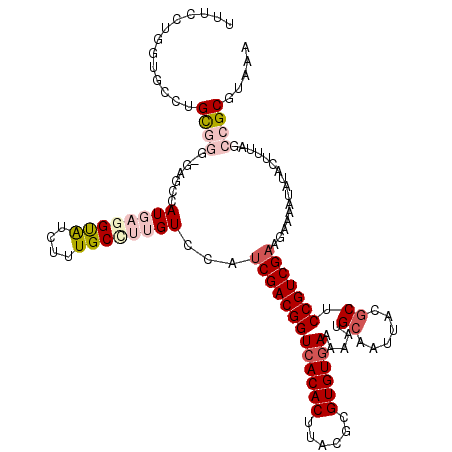
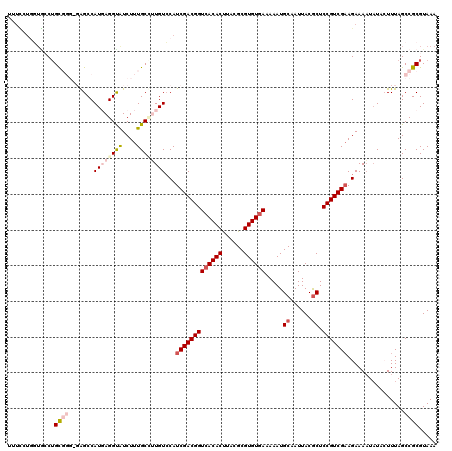
>3R_DroMel_CAF1 2472574 119 + 27905053 UUUUCUGGUGCCAGUGGG-GAGCCAUGAGGCAUCUUUGCUUUGUCCAUCGACGGUCACACUUACGCGUGUGAAAAAUGCAAUUACACUCCGUCGAAGAAAAUAUACUUUAGCCGCGUAAA ((((((((((((.((((.-...))))..)))))).............(((((((((((((......))))))....((......))..)))))))))))))................... ( -35.80) >DroSec_CAF1 2706 104 + 1 -----------CUGC-----GGCCAUGAGGUAUCUUUGCCUUGUCCAUCGACGGUCACACUUACGCGUGUGAAAAAUGCAAUUACGCUCCGUCGAAGAAAAUAUACUUUAGCCGCGUAAA -----------..((-----(((.(..(((((....)))))..)...(((((((((((((......)))))).....((......)).)))))))...............)))))..... ( -37.40) >DroSim_CAF1 4570 115 + 1 UUUUCUUGUAGCUGC-----GGCCAUGAGGUAUCUUUGCCUUGUCCAUCGACGGUCACACUUACGCGUGUGAAAAAUGCAAUUACGCUCCGUCGAAGAAAAUAUACUUUAGCCGCGUAAA .............((-----(((.(..(((((....)))))..)...(((((((((((((......)))))).....((......)).)))))))...............)))))..... ( -37.40) >DroEre_CAF1 2830 118 + 1 UUUCUUGUUGCCUGCGGGGGAGCCAUUAGGUGUCUUUGCGAUGUACAUCGACGGUCACACUAACGCGUGUGAUAAAUGCAAUUACGCUCCGUCGUAGAAAUUAU--UUUAGCCGCGUAAA ......((.((((((((.(((((..(((((((.((...((((....))))..)).))).)))).((((.......))))......))))).))))))(((....--))).)).))..... ( -34.90) >DroYak_CAF1 4044 118 + 1 UUUCUUGGUGCCUGCGGGAGAGCCAUUAGGUAUCUUUGCGAUGUCCAUCGACGGUCACACCCACGCGUGUAAUAAAUGCAAUUACGCUCCGUCGAAGAAAAUUU--UUUAGCCGCGUAAA ......((((((((..((....))..)))))))).((((((((((....))).)))((((......)))).......))))((((((...((..((((....))--))..)).)))))). ( -29.50) >consensus UUUCCUGGUGCCUGCGGG_GAGCCAUGAGGUAUCUUUGCCUUGUCCAUCGACGGUCACACUUACGCGUGUGAAAAAUGCAAUUACGCUCCGUCGAAGAAAAUAUACUUUAGCCGCGUAAA .............((((.......((((((((....))))))))...(((((((((((((......)))))).....((......)).)))))))................))))..... (-23.34 = -25.38 + 2.04)

| Location | 2,472,574 – 2,472,693 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.40 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
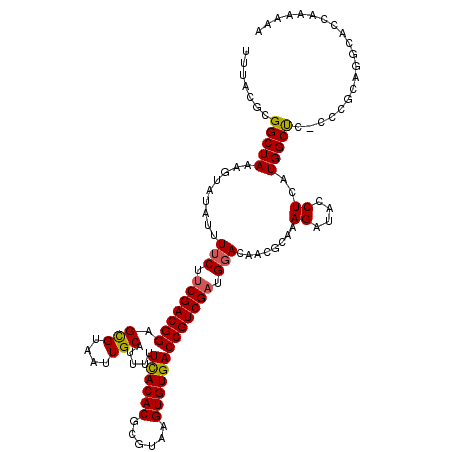
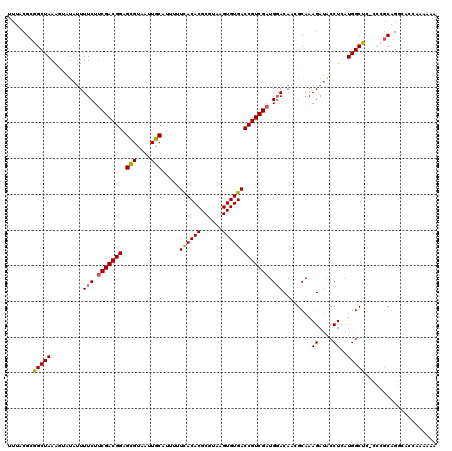
>3R_DroMel_CAF1 2472574 119 - 27905053 UUUACGCGGCUAAAGUAUAUUUUCUUCGACGGAGUGUAAUUGCAUUUUUCACACGCGUAAGUGUGACCGUCGAUGGACAAAGCAAAGAUGCCUCAUGGCUC-CCCACUGGCACCAGAAAA .....((.((....)).....(((.((((((((((((....))))).....(((((....))))).))))))).)))....))...((.(((....)))))-....(((....))).... ( -33.70) >DroSec_CAF1 2706 104 - 1 UUUACGCGGCUAAAGUAUAUUUUCUUCGACGGAGCGUAAUUGCAUUUUUCACACGCGUAAGUGUGACCGUCGAUGGACAAGGCAAAGAUACCUCAUGGCC-----GCAG----------- .....(((((((.........(((.(((((((.(((....)))........(((((....))))).))))))).)))..(((........)))..)))))-----))..----------- ( -39.00) >DroSim_CAF1 4570 115 - 1 UUUACGCGGCUAAAGUAUAUUUUCUUCGACGGAGCGUAAUUGCAUUUUUCACACGCGUAAGUGUGACCGUCGAUGGACAAGGCAAAGAUACCUCAUGGCC-----GCAGCUACAAGAAAA .....(((((((.........(((.(((((((.(((....)))........(((((....))))).))))))).)))..(((........)))..)))))-----))............. ( -39.00) >DroEre_CAF1 2830 118 - 1 UUUACGCGGCUAAA--AUAAUUUCUACGACGGAGCGUAAUUGCAUUUAUCACACGCGUUAGUGUGACCGUCGAUGUACAUCGCAAAGACACCUAAUGGCUCCCCCGCAGGCAACAAGAAA .....((((.....--......(((.((((((.(((....))).....((((((......)))))))))))).(((.....))).)))..((....)).....)))).(....)...... ( -31.10) >DroYak_CAF1 4044 118 - 1 UUUACGCGGCUAAA--AAAUUUUCUUCGACGGAGCGUAAUUGCAUUUAUUACACGCGUGGGUGUGACCGUCGAUGGACAUCGCAAAGAUACCUAAUGGCUCUCCCGCAGGCACCAAGAAA .....((((.....--.....(((.(((((((.(((....)))........(((((....))))).))))))).))).(((.....))).((....)).....))))............. ( -30.00) >consensus UUUACGCGGCUAAAGUAUAUUUUCUUCGACGGAGCGUAAUUGCAUUUUUCACACGCGUAAGUGUGACCGUCGAUGGACAACGCAAAGAUACCUCAUGGCUC_CCCGCAGGCACCAAAAAA .......(((((.........(((.(((((((.(((....))).....((((((......))))))))))))).)))........((....))..))))).................... (-25.56 = -25.40 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:38 2006