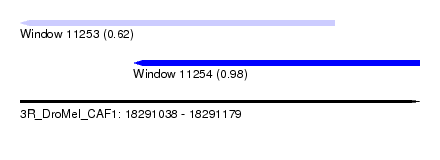
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,291,038 – 18,291,179 |
| Length | 141 |
| Max. P | 0.977857 |

| Location | 18,291,038 – 18,291,149 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -25.15 |
| Energy contribution | -26.90 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18291038 111 - 27905053 UCCCUCCAGCCCAGACCAGCUGACAAUCGGUUGGUCCUUCACCUGGGUGGGGGUUAGCUCGUUGCCCUGCUUGACCUCCAAGUCAUCGCCAUAGAUGACCUUAAUGGUGCC ...(.(((((((((((((((((.....))))))))).......)))))((((((((((..........)).))))))))..((((((......)))))).....))).).. ( -43.31) >DroVir_CAF1 2518 111 - 1 GCCCUCCGCGGCCUCCCAGGUCACAAGCGGCUCGUCCUUUACCUGGGUGGGCGUCAGCUCGUUGCCCUCCUGGACAGCGAGCUCACCCGCAUAGCUGACCUUCAGCUUGUC .......((((((((((((((...(((.((....))))).))))))).))))((.(((((((((.((....)).))))))))).)).)))..(((((.....))))).... ( -49.60) >DroSec_CAF1 2728 111 - 1 UCCCUCCAGCCCAGCCCAGCUGACAAUCGGCUGGUCCUUCACCUGGGUGGGGGUCAGCUCGUUGCCCUGCUUGACCUCCAGGUCAUCGCCAUAGAUGACCUUAACGGUAUC .....((.((((((.(((((((.....)))))))........))))))((((((((((..........)).))))))))((((((((......))))))))....)).... ( -48.40) >DroSim_CAF1 2726 111 - 1 UCCCUCCAGCCCAGACCAGCUGACAAUCGGUUGAUCCUUCACCUGAGUGGGGGUCAGCUCGUUGCCCUGCUUCACCUCCAGGUCAUCGACAUAGAUGACCGUUAUGGUAUC ......((((........)))).((((.((((((((((.(((....))))))))))))).)))).........(((....(((((((......))))))).....)))... ( -38.60) >DroEre_CAF1 2694 111 - 1 UCCCUCCAGCCCAGACCAGCUGACAAUGGGUUGAUCCUUCACCUGGGUGGGGGUCAGCUCGUUGCCCUGCUUCAGAUCGAGGCCAUCGCCAUAGACGACCUUUAUCGUGCC ......((((........)))).(((((((((((((((.(((....))))))))))))))))))....((....(((.((((.(.((......)).).)))).)))..)). ( -39.10) >DroYak_CAF1 2726 111 - 1 UCCCUCCAGACCAGACCAGCUGACAUUCGGUUGAUCCUUCACCUGGGUGGGGGUCAGCUCGUUACCUUGCUUCACCUCCAAGCUAUCGCCAUAGGCGACCUUUAUGGUGCC .............((.((((((.....)))))).))...((((.((((((((((.(((..........)))..))))))).....(((((...))))))))....)))).. ( -33.60) >consensus UCCCUCCAGCCCAGACCAGCUGACAAUCGGUUGAUCCUUCACCUGGGUGGGGGUCAGCUCGUUGCCCUGCUUCACCUCCAGGUCAUCGCCAUAGAUGACCUUUAUGGUGCC .................(((...((((.((((((.(((((((....))))))))))))).))))....)))..(((...((((((((......))))))))....)))... (-25.15 = -26.90 + 1.75)



| Location | 18,291,078 – 18,291,179 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18291078 101 - 27905053 CACCAUGAGCAGGGUCAAUAGGUUGGAC---UUUCCCUCCAGC---------CCAGACCAGCUGACAAUCGGUUGGUCCUUCACCUGGGUGGGGGUUAGCUCGUUGCCCUGCU .......(((((((.(((..((((((..---....((((((.(---------((((((((((((.....))))))))).......))))))))))))))))..)))))))))) ( -48.11) >DroVir_CAF1 2558 104 - 1 CACCAUCAACAGCGUAUGCAGAGUUGCUUCAUCGCCCUCCGCG---------GCCUCCCAGGUCACAAGCGGCUCGUCCUUUACCUGGGUGGGCGUCAGCUCGUUGCCCUCCU ......((((.((....)).(((((((.....(((.....)))---------(((((((((((...(((.((....))))).))))))).))))).))))))))))....... ( -34.80) >DroPse_CAF1 2965 107 - 1 GACCAUGAGUAGGGUGAGCAGGGUA------UCGCGCUCCUGCCCCUCAUCGCCUUCCCAGGUCACUUUCGGCUCAUCCUUCACCUGUGUGGGGGUCAGCGCGUUGCCCUGCU .(((........))).(((((((((------.((((((...(((.......((((....)))).......)))....(((((((....)))))))..)))))).))))))))) ( -43.94) >DroSec_CAF1 2768 101 - 1 CACCAUGAGCAAGGUCAAUAGGUUAGAC---UUUCCCUCCAGC---------CCAGCCCAGCUGACAAUCGGCUGGUCCUUCACCUGGGUGGGGGUCAGCUCGUUGCCCUGCU .......((((.((.(((..((((.(((---(..((((((((.---------..((.(((((((.....)))))))..))....))))).)))))))))))..))))).)))) ( -40.90) >DroEre_CAF1 2734 101 - 1 CACCAUGAGCAGGGUGAAUAGGUUGGAC---UUUCCCUCCAGC---------CCAGACCAGCUGACAAUGGGUUGAUCCUUCACCUGGGUGGGGGUCAGCUCGUUGCCCUGCU .......(((((((......(((((((.---......))))))---------)(((.....))).(((((((((((((((.(((....))))))))))))))))))))))))) ( -48.30) >DroPer_CAF1 2982 107 - 1 CACCAUGAGUAGGGUGAGCAGCGUA------UCGCGCUCCUGCCCCUCAUCGCUUUCCCAGGUCACUUUCGGCUCAUCCUUCACCUGUGUGGGGGUCAGCGCGUUGCCCUGCU ((((........))))(((((.(((------.((((((...(((((.((.(((......(((...............)))......)))))))))).)))))).))).))))) ( -39.26) >consensus CACCAUGAGCAGGGUGAACAGGGUAGAC___UCGCCCUCCAGC_________CCAGCCCAGCUCACAAUCGGCUCAUCCUUCACCUGGGUGGGGGUCAGCUCGUUGCCCUGCU .......((((((((.(((.(((.................................)))...........((((...(((((((....)))))))..))).)))))))))))) (-20.30 = -20.44 + 0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:41 2006