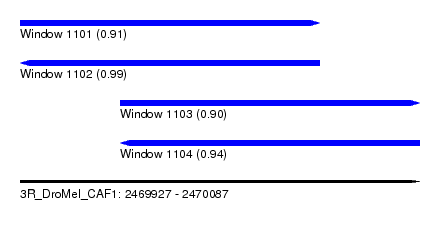
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,469,927 – 2,470,087 |
| Length | 160 |
| Max. P | 0.987187 |

| Location | 2,469,927 – 2,470,047 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2469927 120 + 27905053 GUUGGACAACCUUUUUUGCGCCAAUCUUAAGAGCCUUCUUCUCGGCGGCGGUGAAAUCUUCCUUCUGGCCUACAUCGGCCAGGACGCAAAUGACCUCAUACUGCUUGUCCAGCAACCACU (((((((((....(((..((((..((....))(((........)))))).)..)))......((((((((......)))))))).(((.(((....)))..))))))))))))....... ( -41.20) >DroVir_CAF1 23181 117 + 1 --UGG-AGACCUCCUUGGCGCCUAUCUUGAGCGCCUUUUGACGAGCUGCCUCAAAGUCCUCCUGCUGACCCACAUCGGCCAGCACGCAGAUGACCUCAUAGCGCUGCUCCAGCAGCCACU --.((-((..((....((((((......).))))).......(((....)))..))..))))(((((.((......)).))))).((..(((....))).))(((((....))))).... ( -36.90) >DroPse_CAF1 28720 120 + 1 GUUAUCUCACCUUAUUGGCGCCGAUAUUGAGAGCCUUCUUUUCGGCCGCUGCAAAGUCCUCUUUCUGGCCAACAUCAGCCAAGACACAGAUGACGUUGUACUGCUGGUCCAGUAGCCAUU (((((((.......((((((((((....(((......))).))))).((((.((((....)))).))))........))))).....)))))))......((((((...))))))..... ( -33.60) >DroMoj_CAF1 14403 118 + 1 --UAACCCACCUCCUUGGCUCCGAUUUUGAGCGCCUUCUGGCGGGCUGCCUCAAAGUUCUCCUGCUGGCCAACAUCGGCCAGCACACAGAUAACCUCAUAGCGCUGCUCCAGCAGCCAUU --...(((.........((((.......))))(((....))))))..((......(((.((.((((((((......))))))))....)).)))......))(((((....))))).... ( -40.70) >DroAna_CAF1 10718 113 + 1 -------UACCUUCUUGGCUCCUAUUUUCAGAGCCUUUUUCUCGGCCGCCACAAAGUCCUCUUUUUGGCCCACAUCAGCCAGAACACAGAUUACCUCGUACUUUUGGUCCAGCAACCACU -------.........(((((.........)))))........((..((((.((((....)))).))))........(((((((....((.....))....))))))).......))... ( -22.40) >DroPer_CAF1 27512 120 + 1 GUUAUCUCACCUUAUUGGCGCCGAUAUUGAGAGCCUUCUUUUCGGCCGCUACAAAGUCCUCUUUCUGGCCAACAUCAGCCAAGACACAGAUGACGUUGUACUGCUGGUCCAGUAGCCAUU (((((((.......((((((((((....(((......))).))))).((((.((((....)))).))))........))))).....)))))))......((((((...))))))..... ( -34.90) >consensus __UAGCCCACCUUCUUGGCGCCGAUAUUGAGAGCCUUCUUCUCGGCCGCCACAAAGUCCUCCUUCUGGCCAACAUCAGCCAGGACACAGAUGACCUCAUACUGCUGGUCCAGCAGCCACU ...............((((.............(((........))).........(((.((.(((((((........)))))))....)).)))........((((...)))).)))).. (-16.09 = -16.95 + 0.86)

| Location | 2,469,927 – 2,470,047 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.40 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -30.45 |
| Energy contribution | -28.98 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2469927 120 - 27905053 AGUGGUUGCUGGACAAGCAGUAUGAGGUCAUUUGCGUCCUGGCCGAUGUAGGCCAGAAGGAAGAUUUCACCGCCGCCGAGAAGAAGGCUCUUAAGAUUGGCGCAAAAAAGGUUGUCCAAC .........(((((((.(....(((((((.(((...((.(((((......))))))).))).)))))))..((.(((((.((((....))))....)))))))......).))))))).. ( -41.00) >DroVir_CAF1 23181 117 - 1 AGUGGCUGCUGGAGCAGCGCUAUGAGGUCAUCUGCGUGCUGGCCGAUGUGGGUCAGCAGGAGGACUUUGAGGCAGCUCGUCAAAAGGCGCUCAAGAUAGGCGCCAAGGAGGUCU-CCA-- (((.(((((....)))))(((.(((((((.....(.((((((((......)))))))).)..))))))).))).)))........((((((.......))))))..(((....)-)).-- ( -49.70) >DroPse_CAF1 28720 120 - 1 AAUGGCUACUGGACCAGCAGUACAACGUCAUCUGUGUCUUGGCUGAUGUUGGCCAGAAAGAGGACUUUGCAGCGGCCGAAAAGAAGGCUCUCAAUAUCGGCGCCAAUAAGGUGAGAUAAC ..(((((((((......)))))((((((((.(((.....))).))))))))))))...........(((.((.((((........)))))))))((((..((((.....)))).)))).. ( -35.50) >DroMoj_CAF1 14403 118 - 1 AAUGGCUGCUGGAGCAGCGCUAUGAGGUUAUCUGUGUGCUGGCCGAUGUUGGCCAGCAGGAGAACUUUGAGGCAGCCCGCCAGAAGGCGCUCAAAAUCGGAGCCAAGGAGGUGGGUUA-- ....(((((....)))))(((.(((((((.(((...(((((((((....)))))))))))).))))))).)))((((((((.....(.((((.......))))).....)))))))).-- ( -54.70) >DroAna_CAF1 10718 113 - 1 AGUGGUUGCUGGACCAAAAGUACGAGGUAAUCUGUGUUCUGGCUGAUGUGGGCCAAAAAGAGGACUUUGUGGCGGCCGAGAAAAAGGCUCUGAAAAUAGGAGCCAAGAAGGUA------- ..((((((((..((.....))(((((((..(((...((.(((((......))))))).)))..))))))))))))))).......((((((.......)))))).........------- ( -35.30) >DroPer_CAF1 27512 120 - 1 AAUGGCUACUGGACCAGCAGUACAACGUCAUCUGUGUCUUGGCUGAUGUUGGCCAGAAAGAGGACUUUGUAGCGGCCGAAAAGAAGGCUCUCAAUAUCGGCGCCAAUAAGGUGAGAUAAC .((((((((((......)))))....)))))...((((((.((((((((((((.(.((((....)))).).))((((........))))..))))))))))(((.....))))))))).. ( -36.80) >consensus AAUGGCUGCUGGACCAGCAGUACGAGGUCAUCUGUGUCCUGGCCGAUGUUGGCCAGAAAGAGGACUUUGAGGCGGCCGAAAAGAAGGCUCUCAAAAUCGGCGCCAAGAAGGUGAGAUA__ ...((((((((.((.....)).(((((((.(((.....((((((......))))))..))).)))))))))))))))........(((.((.......)).)))................ (-30.45 = -28.98 + -1.47)

| Location | 2,469,967 – 2,470,087 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -20.29 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2469967 120 + 27905053 CUCGGCGGCGGUGAAAUCUUCCUUCUGGCCUACAUCGGCCAGGACGCAAAUGACCUCAUACUGCUUGUCCAGCAACCACUUGAGGACGCAACUAGUGUCCAGGCCACCGGAGUAAGCCAA .((((.((((((.............(((((......)))))(((((((.(((....)))..)))..))))....)))......((((((.....))))))..))).)))).......... ( -41.80) >DroVir_CAF1 23218 120 + 1 ACGAGCUGCCUCAAAGUCCUCCUGCUGACCCACAUCGGCCAGCACGCAGAUGACCUCAUAGCGCUGCUCCAGCAGCCACUUUAGUAUGCAGCUCGUAUCCAGGCCACCCGAAUAGGCCAA (((((((((..((((((.....(((((.((......)).))))).((..(((....))).))(((((....))))).))))).)...))))))))).....((((.........)))).. ( -44.10) >DroPse_CAF1 28760 120 + 1 UUCGGCCGCUGCAAAGUCCUCUUUCUGGCCAACAUCAGCCAAGACACAGAUGACGUUGUACUGCUGGUCCAGUAGCCAUUUAAGAAUGCAGCUGGUGUCCAAUCCGCCGGAAUAGGCCAA ...((((((((((..(((.(((.((((((........))).)))...))).)))......((((((...))))))...........))))))(((((.......))))).....)))).. ( -39.70) >DroGri_CAF1 15192 120 + 1 UCUUGCUGCCUCAAAGUUCUCCUGCUGACCGACAUCGGCCAGCACGCAGAUGACCUCGUAGCGCUGCUCCAGCAACCACUUGAGUAUGCAGCUUGUGUCGAGGCCACCGGAGUAGGCCAA .(((((((((((.....((((.(((((.(((....))).))))).).))).(((..((.((((((((((.((......)).))))).)).))))).))))))))......)))))).... ( -40.80) >DroMoj_CAF1 14441 120 + 1 GCGGGCUGCCUCAAAGUUCUCCUGCUGGCCAACAUCGGCCAGCACACAGAUAACCUCAUAGCGCUGCUCCAGCAGCCAUUUGAGUAUGCAGCUCGUAUCCAGGCCACCCGAAUAGGCCAA (((((((((((((((..(((..((((((((......))))))))...)))..........(.(((((....)))))).))))))...))))))))).....((((.........)))).. ( -53.30) >DroAna_CAF1 10751 120 + 1 CUCGGCCGCCACAAAGUCCUCUUUUUGGCCCACAUCAGCCAGAACACAGAUUACCUCGUACUUUUGGUCCAGCAACCACUUGAGGAUACAGCUCGUAUCCAGGCCCCCAGAGUAGGCUAA ...((((.......((((....(((((((........)))))))....))))......((((((.((.((..(((....))).((((((.....)))))).))..)).)))))))))).. ( -31.10) >consensus CCCGGCUGCCUCAAAGUCCUCCUGCUGGCCAACAUCGGCCAGCACACAGAUGACCUCAUACCGCUGCUCCAGCAACCACUUGAGUAUGCAGCUCGUAUCCAGGCCACCGGAAUAGGCCAA ...(((.((......(((.((.((((((((......))))))))....)).)))((((....((((......))))....))))...)).)))........((((.........)))).. (-20.29 = -21.72 + 1.42)

| Location | 2,469,967 – 2,470,087 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -48.12 |
| Consensus MFE | -34.13 |
| Energy contribution | -33.00 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2469967 120 - 27905053 UUGGCUUACUCCGGUGGCCUGGACACUAGUUGCGUCCUCAAGUGGUUGCUGGACAAGCAGUAUGAGGUCAUUUGCGUCCUGGCCGAUGUAGGCCAGAAGGAAGAUUUCACCGCCGCCGAG (((((....(((((..(((.((((.(.....).))))......)))..)))))...((.((..((((((.(((...((.(((((......))))))).))).)))))))).)).))))). ( -45.60) >DroVir_CAF1 23218 120 - 1 UUGGCCUAUUCGGGUGGCCUGGAUACGAGCUGCAUACUAAAGUGGCUGCUGGAGCAGCGCUAUGAGGUCAUCUGCGUGCUGGCCGAUGUGGGUCAGCAGGAGGACUUUGAGGCAGCUCGU ..((((.........)))).....(((((((((.......((((.((((....))))))))..((((((.....(.((((((((......)))))))).)..))))))...))))))))) ( -55.20) >DroPse_CAF1 28760 120 - 1 UUGGCCUAUUCCGGCGGAUUGGACACCAGCUGCAUUCUUAAAUGGCUACUGGACCAGCAGUACAACGUCAUCUGUGUCUUGGCUGAUGUUGGCCAGAAAGAGGACUUUGCAGCGGCCGAA ((((((...(((((....))))).....((((((.((((...(((((((((......)))))((((((((.(((.....))).)))))))))))).....))))...)))))))))))). ( -46.10) >DroGri_CAF1 15192 120 - 1 UUGGCCUACUCCGGUGGCCUCGACACAAGCUGCAUACUCAAGUGGUUGCUGGAGCAGCGCUACGAGGUCAUCUGCGUGCUGGCCGAUGUCGGUCAGCAGGAGAACUUUGAGGCAGCAAGA ...((((.((((((((((((((......(((((....((.(((....))).)))))))....))))))))))....(((((((((....))))))))))))).......))))....... ( -53.30) >DroMoj_CAF1 14441 120 - 1 UUGGCCUAUUCGGGUGGCCUGGAUACGAGCUGCAUACUCAAAUGGCUGCUGGAGCAGCGCUAUGAGGUUAUCUGUGUGCUGGCCGAUGUUGGCCAGCAGGAGAACUUUGAGGCAGCCCGC ..((((.........))))......((.(((((...(((.....(((((....))))).....((((((.(((...(((((((((....)))))))))))).)))))))))))))).)). ( -49.90) >DroAna_CAF1 10751 120 - 1 UUAGCCUACUCUGGGGGCCUGGAUACGAGCUGUAUCCUCAAGUGGUUGCUGGACCAAAAGUACGAGGUAAUCUGUGUUCUGGCUGAUGUGGGCCAAAAAGAGGACUUUGUGGCGGCCGAG (..(((...((..(.((((.((((((.....))))))......)))).)..))......(((((((((..(((...((.(((((......))))))).)))..))))))).)))))..). ( -38.60) >consensus UUGGCCUACUCCGGUGGCCUGGACACGAGCUGCAUACUCAAGUGGCUGCUGGACCAGCACUACGAGGUCAUCUGCGUGCUGGCCGAUGUGGGCCAGAAGGAGGACUUUGAGGCAGCCGAA ..((((.........)))).........(((((...........((((......))))....(((((((.(((....(((((((......))))))).))).)))))))..))))).... (-34.13 = -33.00 + -1.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:36 2006