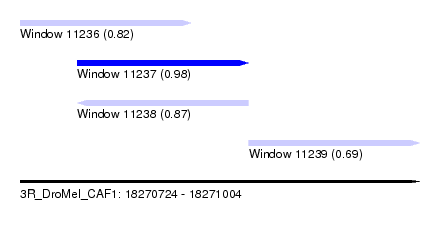
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,270,724 – 18,271,004 |
| Length | 280 |
| Max. P | 0.980631 |

| Location | 18,270,724 – 18,270,844 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -30.15 |
| Energy contribution | -32.02 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18270724 120 + 27905053 GCAUUAAUGAUUUGGGCGAGAAACGCCACAGCUAUGGGCAAAUCUUUCGGCUUGGUUUUGACCGGACUGAGCAGAGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCA ((......((((((((((......(((.((....))))).......((((..((((....))))..))))((((.(((((.....))))).))))....)).)))))))).......)). ( -40.12) >DroSec_CAF1 26943 120 + 1 GCAUUAAUGAUUUGGGCGAGAAACAUCACAGCCUUGGGCAAAUCCGUCGACUUGGUUUUGACCGGACUGAGCAGUGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCA ((......((((((((((..........(((((..((.((((.(((......))).)))).)))).))).((((((((((.....))))))))))....)).)))))))).......)). ( -43.72) >DroSim_CAF1 27930 120 + 1 GCAUUAAUGAUUUGGGCGAGAAACAUAACAGCCUUGGGCAAAUCCGUCGACUUGGUUUUGACCGGACUGAGCAGUGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCA ((......((((((((((..........(((((..((.((((.(((......))).)))).)))).))).((((((((((.....))))))))))....)).)))))))).......)). ( -43.72) >DroYak_CAF1 27443 120 + 1 GCAUCAAUGAUUUGAGCCAGAAAAUUGAAAACCUUUGGCAAAUCCUCCGAUUUGGUUUUGAGCGGACUGAGCAGGGUGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCA ((.((((....))))((((((...(((((((((.((((........))))...))))))....(((((((((((.(.(((.....))).).)))).))).))))))).))))).)..)). ( -33.00) >consensus GCAUUAAUGAUUUGGGCGAGAAACAUCACAGCCUUGGGCAAAUCCGUCGACUUGGUUUUGACCGGACUGAGCAGUGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCA ((......((((((((((..........(((((..((.((((.((........)).)))).)))).))).((((((((((.....))))))))))....)).)))))))).......)). (-30.15 = -32.02 + 1.87)



| Location | 18,270,764 – 18,270,884 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -35.28 |
| Energy contribution | -35.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18270764 120 + 27905053 AAUCUUUCGGCUUGGUUUUGACCGGACUGAGCAGAGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCAGUUAGUGUUUGAAUAACCAAAUCCACUUUGCGCGGAUUAU (((((.((((..((((....))))..))))((((((.(((.........(((((.(((((((((.......))))..))))))))))((((......))))))).))))))..))))).. ( -35.90) >DroSec_CAF1 26983 120 + 1 AAUCCGUCGACUUGGUUUUGACCGGACUGAGCAGUGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCAGUUAGUGUUUUAAUAACCAAAUCCACUUUGCGCGGAUUAU (((((((.((((((((....)))(((((((((((((((((.....)))))))))).))).)))).))))).......((((..((((..((........))..))))))))))))))).. ( -42.80) >DroSim_CAF1 27970 120 + 1 AAUCCGUCGACUUGGUUUUGACCGGACUGAGCAGUGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCAGUUAGUGUUUUAAUAACCAAAUCCACUUUGCGCGGAUUAU (((((((.((((((((....)))(((((((((((((((((.....)))))))))).))).)))).))))).......((((..((((..((........))..))))))))))))))).. ( -42.80) >DroYak_CAF1 27483 120 + 1 AAUCCUCCGAUUUGGUUUUGAGCGGACUGAGCAGGGUGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCAGUUAGUGUUUUUAUGACCAAAUCCACUUUGCGCGGAUUUU (((((((((.(((......)))))))....((((((((((.....((..(((((.(((((((((.......))))..)))))))))).......)).....))))))))))..))))).. ( -35.60) >consensus AAUCCGUCGACUUGGUUUUGACCGGACUGAGCAGUGAGGAACAAUUCCUCACUGUAUUGCGUCCCAAGUCUGGACAAGCAGUUAGUGUUUUAAUAACCAAAUCCACUUUGCGCGGAUUAU ((((((((((..(((.((((....(((((.((((((((((.....)))))))))).....((((.......))))...)))))...(((.....))))))).)))..))).))))))).. (-35.28 = -35.77 + 0.50)



| Location | 18,270,764 – 18,270,884 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -30.14 |
| Energy contribution | -31.45 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18270764 120 - 27905053 AUAAUCCGCGCAAAGUGGAUUUGGUUAUUCAAACACUAACUGCUUGUCCAGACUUGGGACGCAAUACAGUGAGGAAUUGUUCCUCUCUGCUCAGUCCGGUCAAAACCAAGCCGAAAGAUU ..(((((((.....))))))).(((((.........)))))(((((....((((..((((......(((.(((((.....))))).)))....)))))))).....)))))(....)... ( -34.50) >DroSec_CAF1 26983 120 - 1 AUAAUCCGCGCAAAGUGGAUUUGGUUAUUAAAACACUAACUGCUUGUCCAGACUUGGGACGCAAUACAGUGAGGAAUUGUUCCUCACUGCUCAGUCCGGUCAAAACCAAGUCGACGGAUU ..(((((((......(((((..(((..(((......)))..))).)))))(((((.((((......(((((((((.....)))))))))....))))(((....))))))))).)))))) ( -42.10) >DroSim_CAF1 27970 120 - 1 AUAAUCCGCGCAAAGUGGAUUUGGUUAUUAAAACACUAACUGCUUGUCCAGACUUGGGACGCAAUACAGUGAGGAAUUGUUCCUCACUGCUCAGUCCGGUCAAAACCAAGUCGACGGAUU ..(((((((......(((((..(((..(((......)))..))).)))))(((((.((((......(((((((((.....)))))))))....))))(((....))))))))).)))))) ( -42.10) >DroYak_CAF1 27483 120 - 1 AAAAUCCGCGCAAAGUGGAUUUGGUCAUAAAAACACUAACUGCUUGUCCAGACUUGGGACGCAAUACAGUGAGGAAUUGUUCCACCCUGCUCAGUCCGCUCAAAACCAAAUCGGAGGAUU ..(((((......((((((((.(((........((((...(((..((((.......)))))))....)))).(((.....))).....))).)))))))).....((.....)).))))) ( -29.60) >consensus AUAAUCCGCGCAAAGUGGAUUUGGUUAUUAAAACACUAACUGCUUGUCCAGACUUGGGACGCAAUACAGUGAGGAAUUGUUCCUCACUGCUCAGUCCGGUCAAAACCAAGUCGAAGGAUU ..(((((.(((...)))(((((((((.............(((......))).....((((......(((((((((.....)))))))))....))))......)))))))))...))))) (-30.14 = -31.45 + 1.31)



| Location | 18,270,884 – 18,271,004 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -41.95 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18270884 120 + 27905053 CGUAGGGCUUGAAUAGAGAGCCAAGUAAUCUGCCCUGCAGUUUGUUUUGCUUUUGGCGGUAGUGCGUCAUCGCAUCAAAGCAGAUGCCGUGCUUGCGGGAAGUCAAAAGGACUAGCAGGA .(((((((((.......))))).......))))(((((((((..(((.((((((.(((..((..((.((((((......)).)))).))..))))).)))))).)))..)))).))))). ( -44.91) >DroSec_CAF1 27103 120 + 1 CGUAGGGCUUGAAUAGAGAGCCAAGUAAUCUGCCCUGCAGCUUGUUUUGCUUUUGGCGGUAGUGCGUCAUCGCAUCAAAGCAGAUGCCGUGCUUGCGGGAAGUCAGAAGGACCAGCAGGA .(((((((((.......))))).......))))(((((.(.((.((((((((((.(((..((..((.((((((......)).)))).))..))))).))))).))))).)).).))))). ( -42.91) >DroSim_CAF1 28090 120 + 1 CGUAGGGCUUGAAUAGAGAACCAAGUAAUCUGCCCUGCAGCUUGUUUUGCUUUUGGCGGUAGUGCGUCAUCGCAUCAAAGCAGAUGCCGUGCUUGCGGGAAGUCAGAAGGACCAGCAGAA .(((((((.......................))))))).(((.(((((((((((.(((..((..((.((((((......)).)))).))..))))).))))))....))))).))).... ( -40.50) >DroYak_CAF1 27603 120 + 1 UGUACGGCUUGUAUAGAGAGCCAAGUAAUCUGCCCUGCAGUUUGUUUUGCUUUUGGCGGUAGUGCGUCAUCGCAUCAAAGCAGAUGCCGUGCUUGCGGGAAGUAAGUAGGACCAGCAGUA .....(((((.......)))))............((((.((((..(((((((((.(((..((..((.((((((......)).)))).))..))))).)))))))))..))))..)))).. ( -39.50) >consensus CGUAGGGCUUGAAUAGAGAGCCAAGUAAUCUGCCCUGCAGCUUGUUUUGCUUUUGGCGGUAGUGCGUCAUCGCAUCAAAGCAGAUGCCGUGCUUGCGGGAAGUCAGAAGGACCAGCAGGA .(((((((......(((..((...))..)))))))))).(((.(((((((((((.(((..((..((.((((((......)).)))).))..))))).))))))....))))).))).... (-38.29 = -38.10 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:27 2006