
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,265,476 – 18,265,572 |
| Length | 96 |
| Max. P | 0.904058 |

| Location | 18,265,476 – 18,265,572 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.59 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
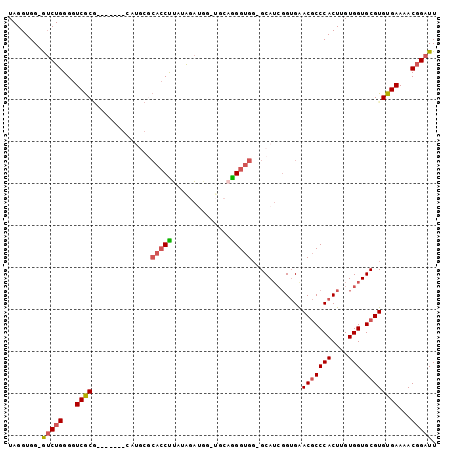
>3R_DroMel_CAF1 18265476 96 + 27905053 UAGGUGG-GUCUGGGGUCGCGAAAGAUGCAUGCACACCUUAUGGA----UGCUGGGUGG-GCAUCGGUGGACGCCCACUUGUGGUGCGUGUGAAAACGGAUU ......(-(((((...((.(....)..((((((((.((....)).----.((.((((((-((.((....)).)))))))))).))))))))))...)))))) ( -36.20) >DroSec_CAF1 21675 100 + 1 UAGGUGG-GUCUGGGGUCGCGAAAGAUGCAUGCGCACCUUAUGGAUGGAUGCAGGGUGG-GCAUCGGUGGACGCCCACUUGUGGUGCGUGUGAAAACGGAUU ......(-(((((...((.(....)..((((((((.((....))......(((((.(((-((.((....)).)))))))))).))))))))))...)))))) ( -36.60) >DroSim_CAF1 22459 100 + 1 UAGGUGG-GUCUGGGGUCGCGAAAGAUGCAUGCGCACCUUAUGGAUGGGUGCAGGGUGG-GCAUCGGUGGACGCCCACUUGUGGUGCGUGUGAAAACGGAUU ......(-(((((...((.(....)..((((((((((((.......)))))).((((((-((.((....)).)))))))).....))))))))...)))))) ( -40.60) >DroEre_CAF1 22317 83 + 1 UAGGUGG-GUCUGGGGUCGC-------------GCACCCUAUAGAUGG----AGGGUGG-GCAUCCGUGAACGCCCACUUGUGGUGCGUGUGAAAACGGAUU (((((((-((...((((.((-------------.((((((........----)))))).-))))))(....)))))))))).(((.(((......))).))) ( -34.80) >DroYak_CAF1 22173 83 + 1 UAGGUGG-AUCUGGGGUCGC-------------GCACCCUAAAGAUGG----AGGGUGG-GCAUCGGUGAACGCCCACUUGUGGUGCGUGUGAAAACGGAUU ......(-(((((...((((-------------.((((((........----)))))).-))........(((((((....))).))))..))...)))))) ( -31.60) >DroAna_CAF1 22801 90 + 1 UAGGUGGCGACUGGGGUCACA-------CAUGAA---CUCGAAGGGGGAUACAAGGCGCUGCA--GGGGAACACCCACUUGUGGUGCGUGUGAAAACCGAUU ...........(((..(((((-------(.....---(((....)))........((((..((--((((.....)).))))..))))))))))...)))... ( -33.50) >consensus UAGGUGG_GUCUGGGGUCGCG_______CAUGCGCACCUUAUAGAUGG_UGCAGGGUGG_GCAUCGGUGAACGCCCACUUGUGGUGCGUGUGAAAACGGAUU ........(((((...((((..............(((((..............)))))............(((((((....))).))))))))...))))). (-20.23 = -20.59 + 0.36)


| Location | 18,265,476 – 18,265,572 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -12.50 |
| Energy contribution | -13.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18265476 96 - 27905053 AAUCCGUUUUCACACGCACCACAAGUGGGCGUCCACCGAUGC-CCACCCAGCA----UCCAUAAGGUGUGCAUGCAUCUUUCGCGACCCCAGAC-CCACCUA .....((((.....(((.......(((((((((....)))))-))))...(((----(.(((.....))).)))).......))).....))))-....... ( -26.80) >DroSec_CAF1 21675 100 - 1 AAUCCGUUUUCACACGCACCACAAGUGGGCGUCCACCGAUGC-CCACCCUGCAUCCAUCCAUAAGGUGCGCAUGCAUCUUUCGCGACCCCAGAC-CCACCUA .....((((.....(((.......(((((((((....)))))-))))..((((((((((.....)))).).)))))......))).....))))-....... ( -28.40) >DroSim_CAF1 22459 100 - 1 AAUCCGUUUUCACACGCACCACAAGUGGGCGUCCACCGAUGC-CCACCCUGCACCCAUCCAUAAGGUGCGCAUGCAUCUUUCGCGACCCCAGAC-CCACCUA .....((((.....(((.......(((((((((....)))))-))))...(((((.........))))).............))).....))))-....... ( -28.30) >DroEre_CAF1 22317 83 - 1 AAUCCGUUUUCACACGCACCACAAGUGGGCGUUCACGGAUGC-CCACCCU----CCAUCUAUAGGGUGC-------------GCGACCCCAGAC-CCACCUA .((((((......((((.(((....)))))))..))))))((-.((((((----........)))))).-------------))..........-....... ( -27.00) >DroYak_CAF1 22173 83 - 1 AAUCCGUUUUCACACGCACCACAAGUGGGCGUUCACCGAUGC-CCACCCU----CCAUCUUUAGGGUGC-------------GCGACCCCAGAU-CCACCUA ............(.((((((....(((((((((....)))))-)))).((----........)))))))-------------).).........-....... ( -24.50) >DroAna_CAF1 22801 90 - 1 AAUCGGUUUUCACACGCACCACAAGUGGGUGUUCCCC--UGCAGCGCCUUGUAUCCCCCUUCGAG---UUCAUG-------UGUGACCCCAGUCGCCACCUA ....((...((((((((.(....((.(((....))))--)...).))((((..........))))---.....)-------)))))..))............ ( -17.80) >consensus AAUCCGUUUUCACACGCACCACAAGUGGGCGUCCACCGAUGC_CCACCCUGCA_CCAUCCAUAAGGUGCGCAUG_______CGCGACCCCAGAC_CCACCUA ..............(((.......(((((((((....))))).))))........((((.....))))..............)))................. (-12.50 = -13.25 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:18 2006