| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,469,274 – 2,469,416 |
| Length | 142 |
| Max. P | 0.929324 |

| Location | 2,469,274 – 2,469,389 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.39 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -12.43 |
| Energy contribution | -11.68 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2469274 115 + 27905053 UUUAAGCCGUAACUUGAAAUGGA---UUUGAUCAGCAUGUCGACGAACCGGUUCUUAAACCGGAACCUACUCGAUCGUUACGGUUUAAACCACAAACCAAAAGCUAAAAUUUUAUGCG-- (((((((((((((((((...((.---.(((((......)))))....((((((....))))))..))...))))..))))))))))))).............((...........)).-- ( -29.80) >DroSim_CAF1 1701 117 + 1 UUUAGGUCGCGGCUUGAAAAGGAACAUUCAAUCAACAUGUCGACGAACCGGUUCUUAAACCGGAACCUUUUCUAUCGGUACGGCUCAAACACCGAACCAA-AGCAAAAAAA--AAGCGCC ....((.(((.(((((((((((.((((.........)))).......((((((....))))))..)))))))....(((.(((........))).))).)-))).......--..))))) ( -31.50) >DroEre_CAF1 1001 117 + 1 UUUAAGCCGUGGUUUGAAAUGGAACCGCGGUGCAGUAUGUCCACGAACCGGUUCCCAAACCGGAACCUUAUCAUUUAAUACGGUACAAACCCCAAACAAAAAGCUAAAAUGAUAUGG--- .....((((((((((......))))))))))................((((((....))))))..((.((((((((.....(((....))).....(.....)...)))))))).))--- ( -30.20) >DroYak_CAF1 1795 98 + 1 UUUAGUCCGUGGCUUGAAAUAAAAACUCGGCUCAGCAUGACGACGAACCGGUUCUCAAAACGGAGCCUUAUCAAUCAAUAUGGCUCGA--------------GCUACAUUGA-------- ........(((((((((...........(((((....(((.(((......))).))).....)))))...(((.......))).))))--------------))))).....-------- ( -24.70) >consensus UUUAAGCCGUGGCUUGAAAUGGAACCUCGGAUCAGCAUGUCGACGAACCGGUUCUCAAACCGGAACCUUAUCAAUCAAUACGGCUCAAACCCCAAACCAA_AGCUAAAAUGA_AUGC___ ....(((((((..((((....................((....))....((((((......)))))).......)))))))))))................................... (-12.43 = -11.68 + -0.75)

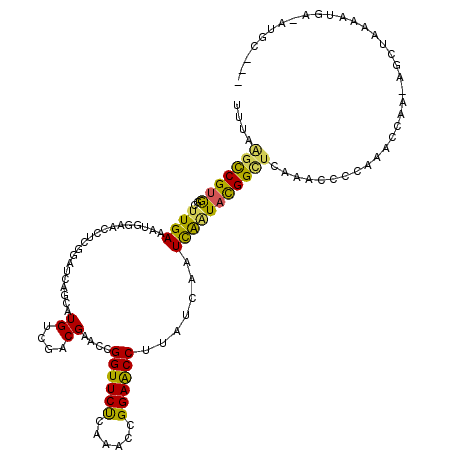
| Location | 2,469,311 – 2,469,416 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 61.93 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -9.20 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2469311 105 + 27905053 CGACGAACCGGUUCUUAAACCGGAACCUACUCGAUCGUUACGGUUUAAACCACAAACCAAAAGCUAAAAUUUUAUGCG--------UUCACGGCACUUCUUGUAGCAUUGAUU .......((((((....)))))).......(((((.((((((((((.......)))))................(((.--------......)))......)))))))))).. ( -21.90) >DroSim_CAF1 1741 110 + 1 CGACGAACCGGUUCUUAAACCGGAACCUUUUCUAUCGGUACGGCUCAAACACCGAACCAA-AGCAAAAAAA--AAGCGCCGUGAACUUCACGGCACUUCUUGUAACAUAGAUU .......((((((....)))))).......(((((.(((.(((........))).)))..-.((((.....--(((.((((((.....)))))).))).))))...))))).. ( -32.30) >DroEre_CAF1 1041 81 + 1 CCACGAACCGGUUCCCAAACCGGAACCUUAUCAUUUAAUACGGUACAAACCCCAAACAAAAAGCUAAAAUGAUAUGG---------------------------GCUC----- .......((((((....))))))..((.((((((((.....(((....))).....(.....)...)))))))).))---------------------------....----- ( -16.30) >consensus CGACGAACCGGUUCUUAAACCGGAACCUUAUCAAUCGAUACGGUUCAAACCCCAAACCAAAAGCUAAAAUA_UAUGCG________UUCACGGCACUUCUUGUAGCAU_GAUU ....(((((((((((......)))))).((......))...)))))................................................................... ( -9.20 = -9.10 + -0.10)

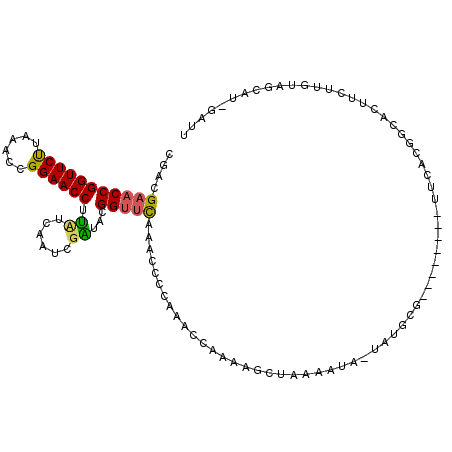
| Location | 2,469,311 – 2,469,416 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.93 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -12.26 |
| Energy contribution | -15.10 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
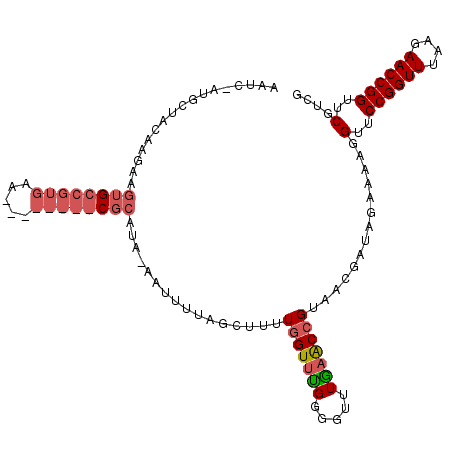
>3R_DroMel_CAF1 2469311 105 - 27905053 AAUCAAUGCUACAAGAAGUGCCGUGAA--------CGCAUAAAAUUUUAGCUUUUGGUUUGUGGUUUAAACCGUAACGAUCGAGUAGGUUCCGGUUUAAGAACCGGUUCGUCG .......((((.((...((((.(....--------)))))....)).))))....((((((.....))))))....(((.((((..(((((........)))))..))))))) ( -25.90) >DroSim_CAF1 1741 110 - 1 AAUCUAUGUUACAAGAAGUGCCGUGAAGUUCACGGCGCUU--UUUUUUUGCU-UUGGUUCGGUGUUUGAGCCGUACCGAUAGAAAAGGUUCCGGUUUAAGAACCGGUUCGUCG ..(((((((...(((((((((((((.....))))))))))--)))....)).-.((((.((((......)))).)))))))))....(..((((((....))))))..).... ( -42.50) >DroEre_CAF1 1041 81 - 1 -----GAGC---------------------------CCAUAUCAUUUUAGCUUUUUGUUUGGGGUUUGUACCGUAUUAAAUGAUAAGGUUCCGGUUUGGGAACCGGUUCGUGG -----((((---------------------------(..((((((((.(((.....)))((.(((....))).))..)))))))).((((((......))))))))))).... ( -25.10) >consensus AAUC_AUGCUACAAGAAGUGCCGUGAA________CGCAUA_AAUUUUAGCUUUUGGUUUGGGGUUUGAACCGUAACGAUAGAAAAGGUUCCGGUUUAAGAACCGGUUCGUCG .................((((((((.....))))))))................(((((((.....)))))))..............(..((((((....))))))..).... (-12.26 = -15.10 + 2.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:32 2006