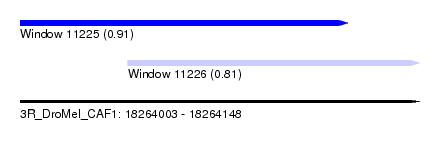
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,264,003 – 18,264,148 |
| Length | 145 |
| Max. P | 0.909692 |

| Location | 18,264,003 – 18,264,122 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -21.52 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18264003 119 + 27905053 AGAAAUUUGUUUAUAAAUAAAUUUGUAUAAAUUGCAUAGAUUUGACAUAAGACAAAGGCCUUUGUGCCCCACGAAAUAAACGUGUCAAUGAGGGCCGCGGUAAUAAUUUAAAAUGCAAA ..(((((((((....))))))))).......((((((...(((((......((...((((((..(....((((.......))))...)..))))))...))......))))))))))). ( -27.60) >DroPse_CAF1 25157 119 + 1 AGAAAUUUGUUUGAAAAUAAAUUUGUAUAAAUUGUAGAUAUUCGAUAUAAGACAAAGGCCUUUGUGCCACAUGAAAUACACAUGUCAAUGAGGGCCGCAGUAAUAAUUUAAAAUGCAAA ..(((((((((....))))))))).......((((..((((...))))...)))).((((((..(...(((((.......)))))..)..))))))(((.(((....)))...)))... ( -25.40) >DroSec_CAF1 20205 119 + 1 AGAAAUUUGUUUAUAAAUAAAUUUGUAUAAAUUGCAUAGAUUUGACAUAAGACAAAGGCCUUUGUGCCCCACAAAAUAAACGUGUCAAUGAGGGCCGCGGUAAUAAUUUAAAAUGCAAA ..(((((((((....))))))))).......((((((...(((((......((...((((((..(....(((.........)))...)..))))))...))......))))))))))). ( -25.10) >DroSim_CAF1 20990 119 + 1 GGAAAUUUGUUUAUAAAUAAAUUUGUAUAAAUUGCAUAGAUUUGACAUAAGACAAAGGCCUUUGUGCCCCACGAAAUAAACGUGUCAAUGAGGGCCGCGGUAAUAAUUUAAAAUGCAAA ..(((((((((....))))))))).......((((((...(((((......((...((((((..(....((((.......))))...)..))))))...))......))))))))))). ( -27.60) >DroAna_CAF1 21468 118 + 1 AGAAAUUUGUUUAAAAAUAAAUUUGUAUAAAUUGCAUACAUUUGACAUAAGACAAUGGCA-UUGUGAGCUAUGAAAUCAACGUGUCAAUGAGGGCCAGACCAAUAAUUUAAAAUGCAAA ..(((((((((....))))))))).......((((((.((((.(((((..((..(((((.-......)))))....))...))))))))).((......))...........)))))). ( -23.00) >DroPer_CAF1 25362 119 + 1 AGAAAUUUGUUUGAAAAUAAAUUUGUAUAAAUUGUAGAUAUUCGAUAUAAGACAAAGGCCUUUGUGCCACAUGAAAUACACAUGUCAAUGAGGGCCGCAGUAAUAAUUUAAAAUGCAAA ..(((((((((....))))))))).......((((..((((...))))...)))).((((((..(...(((((.......)))))..)..))))))(((.(((....)))...)))... ( -25.40) >consensus AGAAAUUUGUUUAAAAAUAAAUUUGUAUAAAUUGCAUAGAUUUGACAUAAGACAAAGGCCUUUGUGCCCCACGAAAUAAACGUGUCAAUGAGGGCCGCAGUAAUAAUUUAAAAUGCAAA (.(((((((((....))))))))).).....((((((...(((((......((...((((((..(....((((.......))))...)..))))))...))......))))))))))). (-21.52 = -22.05 + 0.53)



| Location | 18,264,042 – 18,264,148 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -17.02 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18264042 106 + 27905053 AUUUGACAUAAGACAAAGGCCUUUGUGCCCCACGAAAUAAACGUGUCAAUGAGGGCCGCGGUAAUAAUUUAAAAUGCAAAUAG-----UCGAAA---------AAGGCAAUCGCAGGCCG .((((((..........((((((..(....((((.......))))...)..))))))(((.(((....)))...))).....)-----))))).---------..(((........))). ( -29.80) >DroPse_CAF1 25196 120 + 1 AUUCGAUAUAAGACAAAGGCCUUUGUGCCACAUGAAAUACACAUGUCAAUGAGGGCCGCAGUAAUAAUUUAAAAUGCAAAUAAAUAAACCGAAUAAACUCGACAAGAAAUUCAUGGAUUG (((((............((((((..(...(((((.......)))))..)..))))))(((.(((....)))...)))............))))).......................... ( -24.30) >DroEre_CAF1 20891 106 + 1 AUUUGACAUAAGACAAAGGCCUUUGUGCCCCAUGAAAUAAACGUGUCAAUGAGGGCCGCGGUAAUAAUUUAAAAUGCAAAUAG-----UCGAAA---------AAGGCAAUCGAAGACCG .((((((..........((((((..(....((((.......))))...)..))))))(((.(((....)))...))).....)-----))))).---------..(((.......).)). ( -24.00) >DroYak_CAF1 20758 106 + 1 AUUUGACAUAAGACAAAGGCCUUUGUGCCCCACGAAAUAAACGUGUCAAUGAGGGCCGCGGUAAUAAUUUAAAAUGCAAAUAG-----UCGAAA---------GAGGCAAUCGCAGACCG .((((((..........((((((..(....((((.......))))...)..))))))(((.(((....)))...))).....)-----))))).---------...((....))...... ( -26.40) >DroAna_CAF1 21507 105 + 1 AUUUGACAUAAGACAAUGGCA-UUGUGAGCUAUGAAAUCAACGUGUCAAUGAGGGCCAGACCAAUAAUUUAAAAUGCAAAUAG-----UCGAAA---------AAAGCAAUCGUAGAUUG .((((((......((.(((((-(..(((.........)))..)))))).)).((......))....................)-----))))).---------....(((((...))))) ( -17.60) >DroPer_CAF1 25401 120 + 1 AUUCGAUAUAAGACAAAGGCCUUUGUGCCACAUGAAAUACACAUGUCAAUGAGGGCCGCAGUAAUAAUUUAAAAUGCAAAUAAAUAAACCGAAUAAACUCGACAAGAAAUUCAUGGAUUG (((((............((((((..(...(((((.......)))))..)..))))))(((.(((....)))...)))............))))).......................... ( -24.30) >consensus AUUUGACAUAAGACAAAGGCCUUUGUGCCCCAUGAAAUAAACGUGUCAAUGAGGGCCGCAGUAAUAAUUUAAAAUGCAAAUAG_____UCGAAA_________AAGGCAAUCGUAGACCG .((((............((((((..(....((((.......))))...)..))))))(((.(((....)))...)))............))))........................... (-17.02 = -16.80 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:14 2006