
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,236,626 – 18,236,787 |
| Length | 161 |
| Max. P | 0.993739 |

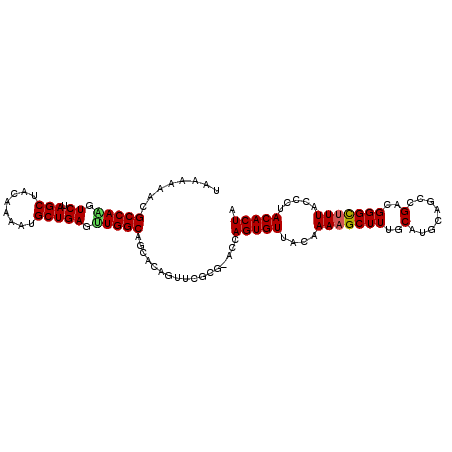
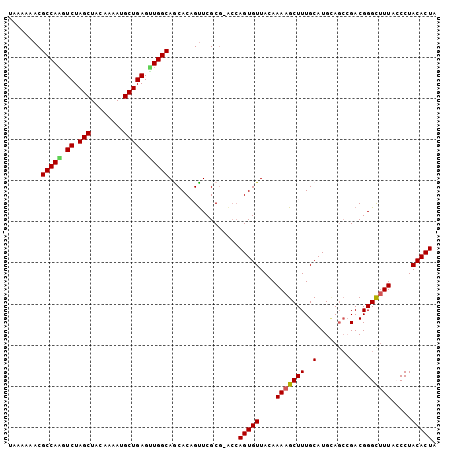
| Location | 18,236,626 – 18,236,725 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.75 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18236626 99 - 27905053 UAAAAAACGCCAGAUCUAGCUAGAAAAUGCUGAGUUGGCAGCACAGCUCGC-----AGUGUUACAAAAGCUUUGCAUGGAGCCGACGGGUCUUACCCUACACUA ........(((((.((.(((........))))).)))))....(.((((((-----((.(((.....))).))))...)))).)..(((.....)))....... ( -25.60) >DroSec_CAF1 4858 103 - 1 UAAAAAACGCCAAGUCUAGCUACAAAAGGCUGAGUUGGCAGCACAGUUCGCG-GCCAGUGUUACAAAAGCUUUGCAUGCAGCCGACGGGCUUUACCCUACACUA ........(((((.((.((((......)))))).))))).....((((((((-((..((((............))))...)))).))))))............. ( -31.30) >DroSim_CAF1 4862 103 - 1 UAAAAAACGCCAAGUCUAGCUACAAAAUGCUGAGUUGGCAGCACAGUUCGCG-ACCAGUGUUACAAAAGCUUUGCAUGCAGCCGACGGGCUUUACCCUACACUA ........(((((.((.(((........))))).))))).((.......)).-...(((((....(((((((.((.....))....))))))).....))))). ( -26.30) >DroEre_CAF1 4869 104 - 1 AGAAAAACGCCAACUCUAGCUACAAAAUGCUGAGCUGGCAACACUGUUCGCGUACCAGUGUUAGAAAAGCUUUGCAUGUAGCAGACGGGCUUUAACCUACACUA ........(((...(((.((((((...(((.(((((..((((((((.........))))))).)...))))).))))))))))))..))).............. ( -33.00) >DroYak_CAF1 4841 100 - 1 A-AUAAACGCCAGGUCUAGCUACAAAAUGCUGAGCUGGCAACACUGUUCGCGUACCAGUGUUACAAAAGCUUUGCAUGUA---GACGGGCUUUAGCCUACACUA .-......((.((((((..(((((...(((.(((((.(.(((((((.........))))))).)...))))).)))))))---)..))))))..))........ ( -30.00) >consensus UAAAAAACGCCAAGUCUAGCUACAAAAUGCUGAGUUGGCAGCACAGUUCGCG_ACCAGUGUUACAAAAGCUUUGCAUGCAGCCGACGGGCUUUACCCUACACUA ........(((((.((.(((........))))).))))).................(((((....(((((((..(........)..))))))).....))))). (-22.62 = -22.58 + -0.04)

| Location | 18,236,661 – 18,236,751 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18236661 90 - 27905053 -------------CAUGC-AAUGACACCUAAGUUCCGGAAUAAAAAACGCCAGAUCUAGCUAGAAAAUGCUGAGUUGGCAGCACAGCUCGC-----AGUGUUACAAAAG -------------.....-..((((((.....................(((((.((.(((........))))).))))).((.......))-----.))))))...... ( -18.70) >DroSec_CAF1 4893 94 - 1 -------------CAUGC-AAUGAUCCGUAAAUAGCGGAUUAAAAAACGCCAAGUCUAGCUACAAAAGGCUGAGUUGGCAGCACAGUUCGCG-GCCAGUGUUACAAAAG -------------..(((-..((((((((.....))))))))......(((((.((.((((......)))))).))))).)))..((.(((.-....)))..))..... ( -30.70) >DroSim_CAF1 4897 95 - 1 -------------CAUGAAAAUGAUCUCUAAGUUGCGGAUUAAAAAACGCCAAGUCUAGCUACAAAAUGCUGAGUUGGCAGCACAGUUCGCG-ACCAGUGUUACAAAAG -------------(((....)))........((((((((((.......(((((.((.(((........))))).))))).....))))))))-)).............. ( -26.40) >DroEre_CAF1 4904 98 - 1 AAAU-----AGUAA------GUAAUCCCUAAUUUCCUGCUAGAAAAACGCCAACUCUAGCUACAAAAUGCUGAGCUGGCAACACUGUUCGCGUACCAGUGUUAGAAAAG ...(-----((((.------(.(((.....))).).))))).......((((.(((.(((........)))))).))))(((((((.........)))))))....... ( -23.00) >DroYak_CAF1 4873 107 - 1 GAUGACGAAAGUACAUGC-UAUAAUCCCUAAGUUCCGGCUA-AUAAACGCCAGGUCUAGCUACAAAAUGCUGAGCUGGCAACACUGUUCGCGUACCAGUGUUACAAAAG .(((((....)).)))((-(..(((......)))..)))..-......(((((.((.(((........))))).)))))(((((((.........)))))))....... ( -28.70) >consensus _____________CAUGC_AAUGAUCCCUAAGUUCCGGAUUAAAAAACGCCAAGUCUAGCUACAAAAUGCUGAGUUGGCAGCACAGUUCGCG_ACCAGUGUUACAAAAG ................................................(((((.((.(((........))))).)))))(((((.............)))))....... (-16.96 = -16.64 + -0.32)

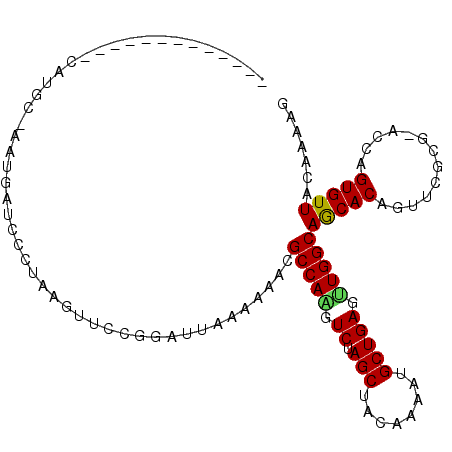
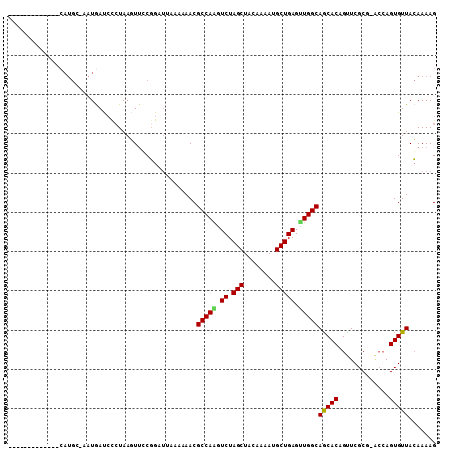
| Location | 18,236,685 – 18,236,787 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -14.02 |
| Energy contribution | -13.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18236685 102 - 27905053 AUAAAAUUGGAAGCAUUUACCAAAUAACCGAAAGCA-----------------CAUGC-AAUGACACCUAAGUUCCGGAAUAAAAAACGCCAGAUCUAGCUAGAAAAUGCUGAGUUGGCA ......((((.........))))....(((...((.-----------------...))-...(((......))).)))..........(((((.((.(((........))))).))))). ( -18.90) >DroSec_CAF1 4921 102 - 1 AUAAAAUUGGAAGCAUUUACCAAAUAACCGAAAGCC-----------------CAUGC-AAUGAUCCGUAAAUAGCGGAUUAAAAAACGCCAAGUCUAGCUACAAAAGGCUGAGUUGGCA ......((((.........))))..........((.-----------------...))-..((((((((.....))))))))......(((((.((.((((......)))))).))))). ( -27.10) >DroSim_CAF1 4925 103 - 1 AUAAAAUUGGAAUCAUUUACCAAAUAUUCGAAAGCC-----------------CAUGAAAAUGAUCUCUAAGUUGCGGAUUAAAAAACGCCAAGUCUAGCUACAAAAUGCUGAGUUGGCA ......((((((((((((..((......(....)..-----------------..)).))))))).))))).................(((((.((.(((........))))).))))). ( -21.80) >DroEre_CAF1 4933 109 - 1 AUGAAAUUGUAAGCAUUUCAUAAAUAACUGAAAGUAAUUUAAAU-----AGUAA------GUAAUCCCUAAUUUCCUGCUAGAAAAACGCCAACUCUAGCUACAAAAUGCUGAGCUGGCA .((((((.(....)))))))(((((.((.....)).)))))..(-----((((.------(.(((.....))).).))))).......((((.(((.(((........)))))).)))). ( -20.40) >DroYak_CAF1 4902 118 - 1 UUGAAAUUUAAAGCAUUUUCCAAAUAACUGAAAGGAAUUUGAUGACGAAAGUACAUGC-UAUAAUCCCUAAGUUCCGGCUA-AUAAACGCCAGGUCUAGCUACAAAAUGCUGAGCUGGCA ...........(((..((((.........))))(((((((((((((....)).)))..-.........)))))))).))).-......(((((.((.(((........))))).))))). ( -28.70) >consensus AUAAAAUUGGAAGCAUUUACCAAAUAACCGAAAGCA_________________CAUGC_AAUGAUCCCUAAGUUCCGGAUUAAAAAACGCCAAGUCUAGCUACAAAAUGCUGAGUUGGCA ........................................................................................(((((.((.(((........))))).))))). (-14.02 = -13.94 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:58 2006