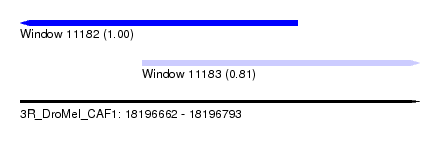
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,196,662 – 18,196,793 |
| Length | 131 |
| Max. P | 0.999464 |

| Location | 18,196,662 – 18,196,753 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -40.70 |
| Energy contribution | -40.58 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18196662 91 - 27905053 UUCACCGCCUGGCGCACAUUGGCCACGGUGGCGGCAUGGUACUCGUGCGAUCCGUGAGAGAAGUUCAUGCGCGCGAUGUUCAUGCCGGUGG ....(((((((((........)))).)))))((((((((.(((((((((...((((((.....))))))))))))).)))))))))).... ( -43.80) >DroSec_CAF1 4738 91 - 1 UUCACCGCCUGGCGCACAUUGGCCACAGUGGCGGCGUGGUACUCGUGCGAUCCGUGAGAGAAGUUCAUGCGCGCGAUGUUCAUGCCGGUGG ..(((((((((((........))))....)))(((((((.(((((((((...((((((.....))))))))))))).))))))))))))). ( -42.80) >DroSim_CAF1 4792 91 - 1 UUCACCGCCUGGCGCACAUUGGCCACGGUGGCGGCGUGGUACUCGUGCGAUCCGUGAGAGAAGUUCAUGCGCGCGAUGUUCAUGCCGGUGG ....(((((((((........)))).)))))((((((((.(((((((((...((((((.....))))))))))))).)))))))))).... ( -43.40) >DroEre_CAF1 4972 91 - 1 UUCACCGCCUGACGCACAUUGGCCACGGUGGCGGCGUGGUACUCGUGCGAUCCGUGAGAGAAGUUCAUGCGCGCAACAUUCAUGCCGGUGG ....(((((....((.(((((....)))))))(((((((.....(((((...((((((.....))))))))))).....)))))))))))) ( -36.70) >DroYak_CAF1 4864 91 - 1 UUAACCGCCUGGCGCACAUUGGCCACGGUGGCGGCGUGGUACUCGUGCGAUCCGUGAGAGAAGUUCAUGCGCGCGACGUUCAUGCCGGUGG ....(((((((((........)))).)))))((((((((.(((((((((...((((((.....))))))))))))).)))))))))).... ( -43.40) >consensus UUCACCGCCUGGCGCACAUUGGCCACGGUGGCGGCGUGGUACUCGUGCGAUCCGUGAGAGAAGUUCAUGCGCGCGAUGUUCAUGCCGGUGG ....(((((....((.(((((....)))))))(((((((...(((((((...((((((.....)))))))))))))...)))))))))))) (-40.70 = -40.58 + -0.12)



| Location | 18,196,702 – 18,196,793 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.24 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18196702 91 + 27905053 AUCGCACGAGUACCAUGCCGCCACCGUGGCCAAUGUGCGCCAGGCGGUGAAGAACUACUCGGCCAAGCUGGGCUACGAACACCCCGUGGCC ...((.((((((......((((.((.(((((.....).)))))).))))......)))))))).......(((((((.......))))))) ( -34.70) >DroSec_CAF1 4778 91 + 1 AUCGCACGAGUACCACGCCGCCACUGUGGCCAAUGUGCGCCAGGCGGUGAAGAACUACUCGGCCAAGCUGGGCUACGAACACCCCGUGGCC ...((.((((((...(((((((....(((((.....).)))))))))))......)))))))).......(((((((.......))))))) ( -36.50) >DroSim_CAF1 4832 91 + 1 AUCGCACGAGUACCACGCCGCCACCGUGGCCAAUGUGCGCCAGGCGGUGAAGAACUACUCGGCCAAGCUGGGCUACGAACACCCCGUGGCC ...((.((((((...(((((((....(((((.....).)))))))))))......)))))))).......(((((((.......))))))) ( -36.50) >DroEre_CAF1 5012 91 + 1 AUCGCACGAGUACCACGCCGCCACCGUGGCCAAUGUGCGUCAGGCGGUGAAGAACUACUCGGCCAAGCUGGGCUACGAACAUCCCGUGGCC ...((.((((((...(((((((....(((((.....).)))))))))))......)))))))).......(((((((.......))))))) ( -33.80) >DroYak_CAF1 4904 91 + 1 AUCGCACGAGUACCACGCCGCCACCGUGGCCAAUGUGCGCCAGGCGGUUAAGAACUACUCGGCCAAGCUGGGCUACGAACACCCCGUGGCC ...((.((((((....((((((....(((((.....).)))))))))).......)))))))).......(((((((.......))))))) ( -34.20) >consensus AUCGCACGAGUACCACGCCGCCACCGUGGCCAAUGUGCGCCAGGCGGUGAAGAACUACUCGGCCAAGCUGGGCUACGAACACCCCGUGGCC ...((.((((((...(((((((....(((((.....).)))))))))))......)))))))).......(((((((.......))))))) (-35.10 = -35.02 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:33 2006