
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,194,882 – 18,195,034 |
| Length | 152 |
| Max. P | 0.955069 |

| Location | 18,194,882 – 18,194,994 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18194882 112 + 27905053 CCUCU-UGGGC-CC-AAACUCUCUCUCCUCUGCGUUUAUGCUCUCUU-UGCUGUCGGGCUACCGACAUUCUGAGCGCUUUU-UCUGCGGCUAGGAAAAUUUCCGCUUGCCAAA-AAAU .....-..(((-..-..........(((((((((.....((.(((..-...((((((....))))))....))).))....-..)))))..))))............)))...-.... ( -28.15) >DroSec_CAF1 2987 113 + 1 CCUCU-UGGGCCCC-AAACUCUCUCUCCUCUUCGUUUGUGCUCUCUU-UUCUGUCGGGCUACCGACAUUCUGAGAGCUUUU-GCUGCGGCUCGGAAAAUUUCCGCUUGCCAAA-AAAU .....-..(((..(-((((..............))))).((((((..-...((((((....))))))....))))))....-)))..(((.((((.....))))...)))...-.... ( -33.24) >DroSim_CAF1 2891 113 + 1 CCUCU-UGGGCCCC-AAACUCUCUCUCCUCUUCGUUUGUGCUCUCUU-UUCUGUCGGGCUACCGACAUUCUGAGCGCUUUU-GCUGCGGCUCGGAAAAUUUCCGCUUGCCAAA-AAAU .....-..(((..(-((((..............))))).((......-...((((((....))))))((((((((((....-...)).)))))))).......))..)))...-.... ( -30.44) >DroEre_CAF1 3191 116 + 1 CCUCUCUGGGC-CC-AAACUCUCUCGCCUCUGCGAUUGUGCUCUCUUUUUCUUUCGGGCUACCGACAUUCUGAGCGCUUUUCGCUGCGGCUCGGGAAAUUUCCGCUUGCCAAAAAAAU ........(((-..-..........(((.(.((((..((((((..........((((....))))......))))))...)))).).))).(((((...)))))...)))........ ( -33.49) >DroYak_CAF1 3105 117 + 1 CCUCUCUGGGC-CCGAAACUCUCUCGCCUCUGCGUUGCUGCUCUCUUUUUCUUUCGGGCUACCGACAUUCUGAGCGCUUUUCGCUGCGGCUCGGGAAAUUUCCGCUUGCCAAAAAAAU ........(((-(.((((.((((..(((.(.(((..((.((((..........((((....))))......))))))....))).).)))..))))..)))).)...)))........ ( -30.09) >consensus CCUCU_UGGGC_CC_AAACUCUCUCUCCUCUGCGUUUGUGCUCUCUU_UUCUGUCGGGCUACCGACAUUCUGAGCGCUUUU_GCUGCGGCUCGGAAAAUUUCCGCUUGCCAAA_AAAU ........(((........................................((((((....))))))((((((((((........)).))))))))...........)))........ (-22.82 = -23.38 + 0.56)

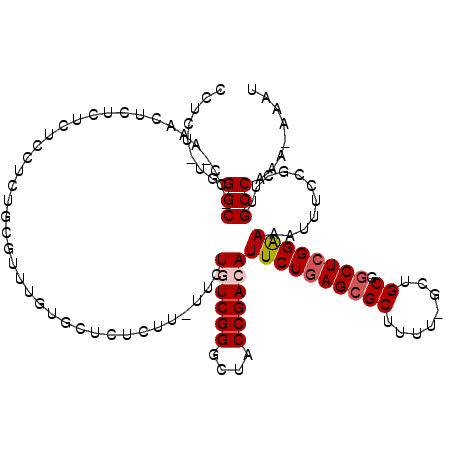

| Location | 18,194,919 – 18,195,034 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18194919 115 + 27905053 CUCUCUU-UGCUGUCGGGCUACCGACAUUCUGAGCGCUUUU-UCUGCGGCUAGGAAAAUUUCCGCUUGCCAAA-AAAUCUGUGUGGCAGUCGAACUUUUUGUCUCGUCCUCGCCGCGU ..(((..-...((((((....))))))....))).......-...(((((.((((..........((((((..-.........)))))).(((((.....)).))))))).))))).. ( -34.70) >DroSec_CAF1 3025 115 + 1 CUCUCUU-UUCUGUCGGGCUACCGACAUUCUGAGAGCUUUU-GCUGCGGCUCGGAAAAUUUCCGCUUGCCAAA-AAAUCUGUGCGGCAGUCGAACUUUUUGUCUCGUCCUCGCCGCGU (((((..-...((((((....))))))....))))).....-.....(((.((((.....))))...)))...-........(((((((.(((((.....)).)))..)).))))).. ( -38.60) >DroSim_CAF1 2929 115 + 1 CUCUCUU-UUCUGUCGGGCUACCGACAUUCUGAGCGCUUUU-GCUGCGGCUCGGAAAAUUUCCGCUUGCCAAA-AAAUCUGUGCGGCAGUCAAACUUUUUGUCUCGUCCUUGCCGCGU ..(((..-...((((((....))))))....))).((....-)).(((((..(((........(((.((((..-.....)).)))))((.(((.....))).))..)))..))))).. ( -33.00) >DroEre_CAF1 3229 118 + 1 CUCUCUUUUUCUUUCGGGCUACCGACAUUCUGAGCGCUUUUCGCUGCGGCUCGGGAAAUUUCCGCUUGCCAAAAAAAUCUGUGCGGCAGUCGAACUUUUUGUCUCGUCCUCGCCGCGU .............((((....))))..((((((((((........)).))))))))..........................(((((((.(((((.....)).)))..)).))))).. ( -31.70) >DroYak_CAF1 3144 118 + 1 CUCUCUUUUUCUUUCGGGCUACCGACAUUCUGAGCGCUUUUCGCUGCGGCUCGGGAAAUUUCCGCUUGCCAAAAAAAUCUGUGCGGCAGUCGAACUUUUUGUCUCGUCCUCGCCGCGU .............((((....))))..((((((((((........)).))))))))..........................(((((((.(((((.....)).)))..)).))))).. ( -31.70) >consensus CUCUCUU_UUCUGUCGGGCUACCGACAUUCUGAGCGCUUUU_GCUGCGGCUCGGAAAAUUUCCGCUUGCCAAA_AAAUCUGUGCGGCAGUCGAACUUUUUGUCUCGUCCUCGCCGCGU ..(((......((((((....))))))....))).((........))(((.(((((...)))))...)))............(((((((.(((((.....)).)))..)).))))).. (-31.20 = -31.60 + 0.40)


| Location | 18,194,919 – 18,195,034 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -30.66 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18194919 115 - 27905053 ACGCGGCGAGGACGAGACAAAAAGUUCGACUGCCACACAGAUUU-UUUGGCAAGCGGAAAUUUUCCUAGCCGCAGA-AAAAGCGCUCAGAAUGUCGGUAGCCCGACAGCA-AAGAGAG ..(((((.((((.((.........((((..(((((.........-..)))))..))))..)).)))).)))))...-.......(((....((((((....))))))...-..))).. ( -36.20) >DroSec_CAF1 3025 115 - 1 ACGCGGCGAGGACGAGACAAAAAGUUCGACUGCCGCACAGAUUU-UUUGGCAAGCGGAAAUUUUCCGAGCCGCAGC-AAAAGCUCUCAGAAUGUCGGUAGCCCGACAGAA-AAGAGAG ..(((((..(((((((........)))).((((.((.((((...-))))))..))))......)))..)))))...-.....(((((....((((((....))))))...-..))))) ( -39.60) >DroSim_CAF1 2929 115 - 1 ACGCGGCAAGGACGAGACAAAAAGUUUGACUGCCGCACAGAUUU-UUUGGCAAGCGGAAAUUUUCCGAGCCGCAGC-AAAAGCGCUCAGAAUGUCGGUAGCCCGACAGAA-AAGAGAG ..(((((..........(((((((((((.........)))))))-)))).....((((.....)))).))))).((-....)).(((....((((((....))))))...-..))).. ( -36.40) >DroEre_CAF1 3229 118 - 1 ACGCGGCGAGGACGAGACAAAAAGUUCGACUGCCGCACAGAUUUUUUUGGCAAGCGGAAAUUUCCCGAGCCGCAGCGAAAAGCGCUCAGAAUGUCGGUAGCCCGAAAGAAAAAGAGAG ..((((((.((((..........))))...)))))).....(((((((((...((.((...(((..((((.((........)))))).)))..)).))...)))))))))........ ( -34.60) >DroYak_CAF1 3144 118 - 1 ACGCGGCGAGGACGAGACAAAAAGUUCGACUGCCGCACAGAUUUUUUUGGCAAGCGGAAAUUUCCCGAGCCGCAGCGAAAAGCGCUCAGAAUGUCGGUAGCCCGAAAGAAAAAGAGAG ..((((((.((((..........))))...)))))).....(((((((((...((.((...(((..((((.((........)))))).)))..)).))...)))))))))........ ( -34.60) >consensus ACGCGGCGAGGACGAGACAAAAAGUUCGACUGCCGCACAGAUUU_UUUGGCAAGCGGAAAUUUUCCGAGCCGCAGC_AAAAGCGCUCAGAAUGUCGGUAGCCCGACAGAA_AAGAGAG ..(((((..((.((((........)))).((((.((.((((....))))))..)))).......))..)))))...........(((....((((((....))))))......))).. (-30.66 = -31.10 + 0.44)

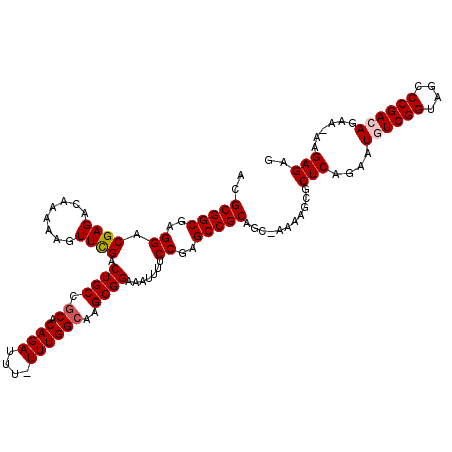

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:28 2006