
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,191,297 – 18,191,388 |
| Length | 91 |
| Max. P | 0.966587 |

| Location | 18,191,297 – 18,191,388 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
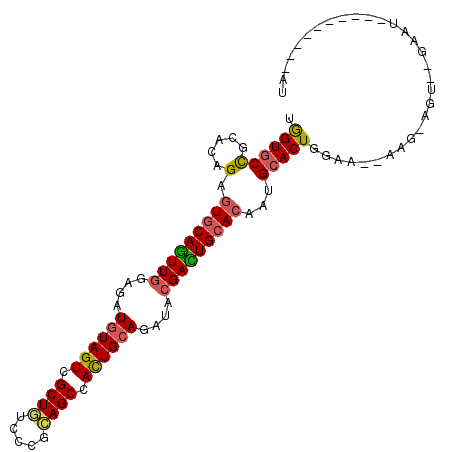
>3R_DroMel_CAF1 18191297 91 + 27905053 UGGUGCCGCACAGAGUGCAGUUGGAGAUGUAGUCGCUGUCCCGCAGCCACUGCAGAUACGACUGCACAAUGCACUGAAA--AAG-AGU--GGAU----------AU .....((((.((..(((((((((....((((((.((((.....)))).))))))....)))))))))..))..((....--.))-.))--))..----------.. ( -36.50) >DroSec_CAF1 4311 91 + 1 UGGUGCCGCACAGAGUGCAGUUGGAGAUGUAGUCGCUGUCCCGCAGCCACUGCAGAUACGACUGCACAAUGCACUGGAA--AAG-AGU--GAAU----------AU .((((((.....).(((((((((....((((((.((((.....)))).))))))....)))))))))...)))))....--...-...--....----------.. ( -35.30) >DroSim_CAF1 4327 91 + 1 UGGUGCCGCACAGAGUGCAGUUGGAGAUGUAGUCGCUGUCCCGCAGCCACUGCAGAUACGACUGCACAAUGCACUGGAA--AAG-AGU--GAAU----------AU .((((((.....).(((((((((....((((((.((((.....)))).))))))....)))))))))...)))))....--...-...--....----------.. ( -35.30) >DroYak_CAF1 4518 92 + 1 UGGUGCUGCACAGAGUGCAGUUGGAGAUGUAGUCGCUGUCCCGCAGCCACUGCAGAUACGACUGCACAAUGCACUGGAA--AACAAAU--UAAU----------GU .(((((........(((((((((....((((((.((((.....)))).))))))....)))))))))...)))))....--.......--....----------.. ( -34.80) >DroMoj_CAF1 4256 101 + 1 CGGUGCCACAAAGCGUACAAUUGGAUAUAUAGUCGCUAUCACGUAGCCAUUGCAGAUACGAUUGCACUAUGCACUGCAA--GAA-AUU--GAUUAACAGUGAAAAU .(((((.....((((.((.((.......)).))))))....((((((....))...))))...)))))...((((((((--...-.))--).....)))))..... ( -19.30) >DroPer_CAF1 4776 95 + 1 AAGUCCCACACAGAUUGCAGUUGGAAAUGUAGUCGCUGUCACGAAGCCACUGCAGAUAGGAUUGCACUAUGCACUGCGGAAUAG-AUCCGAAAU----------AC ..........((...(((((((..(..((((((.(((.......))).))))))..)..)))))))...)).....((((....-.))))....----------.. ( -22.90) >consensus UGGUGCCGCACAGAGUGCAGUUGGAGAUGUAGUCGCUGUCCCGCAGCCACUGCAGAUACGACUGCACAAUGCACUGGAA__AAG_AGU__GAAU__________AU .((((((.....).(((((((((....((((((.((((.....)))).))))))....)))))))))...)))))............................... (-28.56 = -28.57 + 0.00)

| Location | 18,191,297 – 18,191,388 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -21.55 |
| Energy contribution | -21.30 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
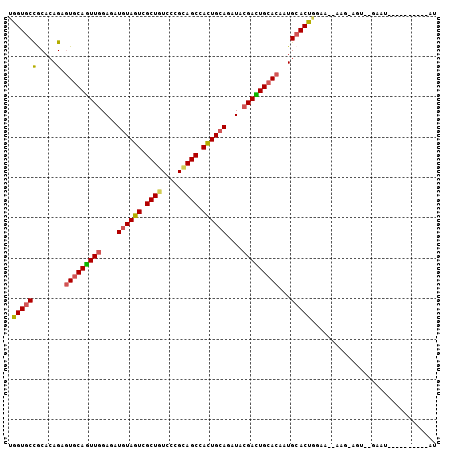
>3R_DroMel_CAF1 18191297 91 - 27905053 AU----------AUCC--ACU-CUU--UUUCAGUGCAUUGUGCAGUCGUAUCUGCAGUGGCUGCGGGACAGCGACUACAUCUCCAACUGCACUCUGUGCGGCACCA ..----------....--...-...--.....(..((..(((((((.(....((.(((.((((.....)))).))).))....).)))))))..))..)....... ( -27.40) >DroSec_CAF1 4311 91 - 1 AU----------AUUC--ACU-CUU--UUCCAGUGCAUUGUGCAGUCGUAUCUGCAGUGGCUGCGGGACAGCGACUACAUCUCCAACUGCACUCUGUGCGGCACCA ..----------....--...-...--..((.(..((..(((((((.(....((.(((.((((.....)))).))).))....).)))))))..))..)))..... ( -27.90) >DroSim_CAF1 4327 91 - 1 AU----------AUUC--ACU-CUU--UUCCAGUGCAUUGUGCAGUCGUAUCUGCAGUGGCUGCGGGACAGCGACUACAUCUCCAACUGCACUCUGUGCGGCACCA ..----------....--...-...--..((.(..((..(((((((.(....((.(((.((((.....)))).))).))....).)))))))..))..)))..... ( -27.90) >DroYak_CAF1 4518 92 - 1 AC----------AUUA--AUUUGUU--UUCCAGUGCAUUGUGCAGUCGUAUCUGCAGUGGCUGCGGGACAGCGACUACAUCUCCAACUGCACUCUGUGCAGCACCA ..----------....--.......--.....(..((..(((((((.(....((.(((.((((.....)))).))).))....).)))))))..))..)....... ( -27.40) >DroMoj_CAF1 4256 101 - 1 AUUUUCACUGUUAAUC--AAU-UUC--UUGCAGUGCAUAGUGCAAUCGUAUCUGCAAUGGCUACGUGAUAGCGACUAUAUAUCCAAUUGUACGCUUUGUGGCACCG .....((((((.....--...-...--..))))))(((..((((........)))))))((((((....((((((.............)).)))).)))))).... ( -21.34) >DroPer_CAF1 4776 95 - 1 GU----------AUUUCGGAU-CUAUUCCGCAGUGCAUAGUGCAAUCCUAUCUGCAGUGGCUUCGUGACAGCGACUACAUUUCCAACUGCAAUCUGUGUGGGACUU ((----------(((.((((.-....)))).))))).........((((((.((((((((..((((....))))........)).))))))......))))))... ( -23.90) >consensus AU__________AUUC__ACU_CUU__UUCCAGUGCAUUGUGCAGUCGUAUCUGCAGUGGCUGCGGGACAGCGACUACAUCUCCAACUGCACUCUGUGCGGCACCA .................................(((((.(((((((......((.(((.((((.....)))).))).))......)))))))...)))))...... (-21.55 = -21.30 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:21 2006