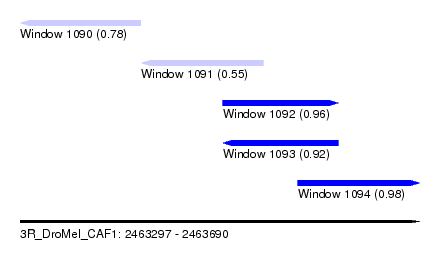
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,463,297 – 2,463,690 |
| Length | 393 |
| Max. P | 0.984964 |

| Location | 2,463,297 – 2,463,416 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.46 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -35.01 |
| Energy contribution | -34.83 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2463297 119 - 27905053 ACAGAAUGUUGCCCUGCCAACGCGGCACGUGGAACGUGGCCAGAUUAUGCUGCAUUAUCACCUGUAAAUCCUUAUUGGCAAUCAAAACUUUGCCAACAGCGCUGGUCCUGACCAUCUCC .(((.(((((.(((((((.....)))).).)))))))((((((....(((((..((((.....)))).......(((((((........)))))))))))))))))))))......... ( -34.00) >DroSec_CAF1 22271 119 - 1 ACAGAAUGUUGCCCUGCCAACGCGGCACGUGGAACGUGGCCAGAUUAUGCUGUAUUAUCACCUGUAAAUCCUUAUUGGCAAUCAAAACUUUGCCAACAGCGCUGGUCCUGACCAGCUCC .(((.(((((.(((((((.....)))).).)))))))((((((....((((((.((((.....))))........((((((........)))))))))))))))))))))......... ( -35.10) >DroSim_CAF1 19011 119 - 1 ACAGAAUGUUGCCCUGCCAACGCGGCACGUGGAACGUGGCCAGAUUAUGCUGUAUUAUCACCUGUAAAUCCUUAUUGGCAAUCAAAACUUUGCCAACAGCGCUGGUCCUGACCAGCUCC .(((.(((((.(((((((.....)))).).)))))))((((((....((((((.((((.....))))........((((((........)))))))))))))))))))))......... ( -35.10) >DroEre_CAF1 23715 119 - 1 ACAGAAUGUUGCCCUGCCAACGCGGCACGUGGAACGUGGCCAGAUUAUGCUGUAUUAUCACCUGUAAAUCCUUAUUGGCAAUCAAAACUUUGCCAACAGCGCUGGCCCUGACCAACUCC .(((.(((((.(((((((.....)))).).)))))))((((((....((((((.((((.....))))........((((((........)))))))))))))))))))))......... ( -37.80) >consensus ACAGAAUGUUGCCCUGCCAACGCGGCACGUGGAACGUGGCCAGAUUAUGCUGUAUUAUCACCUGUAAAUCCUUAUUGGCAAUCAAAACUUUGCCAACAGCGCUGGUCCUGACCAGCUCC .(((.(((((.(((((((.....)))).).)))))))((((((....(((((..((((.....)))).......(((((((........)))))))))))))))))))))......... (-35.01 = -34.83 + -0.19)

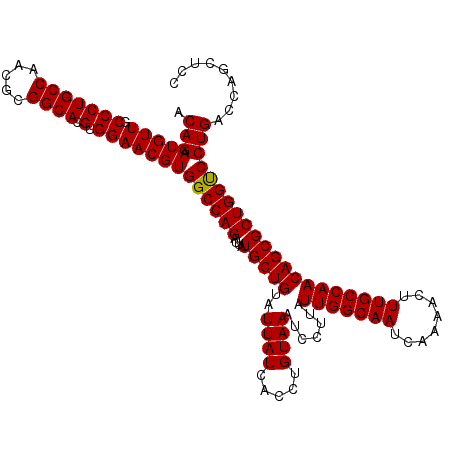
| Location | 2,463,416 – 2,463,536 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -27.20 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2463416 120 - 27905053 UUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUCCGCGCAUGCAGCUCGCAAAAGAAAAGCGGGUAAAAGCCGGGAGCACAAGGUAACUACAAAAUUCCAGAUACAAUUUUACA ....(((((.(((....))).)))))(((((..((((((.((.((.....)).)).))))))....(((.......)))))))).................................... ( -25.30) >DroSec_CAF1 22390 120 - 1 UUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUCCGCGCAUGCAGCUCGCAAAGGAAAAGCGGGUAAAAGCCGGGAGCACAAGGUAACUACAAAAUUCCAGAUACAAUUUUACA ....(((((.(((....))).)))))(((((.(.(((((((.....(((.....)))..))))))).)(((....))).))))).................................... ( -27.20) >DroSim_CAF1 19130 120 - 1 UUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACACUUUUCCGCGGAUGCAGCUCGCAAAGGAAAAGCGGGCAAAAGCCGGGAGCACAAGGUAACUACAAAAUUCCAGAUACAAUUUUACA ....(((((.(((....))).)))))(((((.(.(((((((((((.......))))...))))))).)(((....))).))))).................................... ( -30.10) >DroEre_CAF1 23834 120 - 1 UUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUCCGCGCAUGCAGCUCGCAAAGGAAAAGCGGGCAAAAGCCGGGAGCACAAGGUAACUACAAUUUUCCAGAUACAAUUUUACA ....(((((.(((....))).)))))(((((.(.(((((((.....(((.....)))..))))))).)(((....))).))))).................................... ( -29.10) >consensus UUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUCCGCGCAUGCAGCUCGCAAAGGAAAAGCGGGCAAAAGCCGGGAGCACAAGGUAACUACAAAAUUCCAGAUACAAUUUUACA ....(((((.(((....))).)))))(((((.(.(((((((.....(((.....)))..))))))).)(((....))).))))).................................... (-27.20 = -27.20 + -0.00)

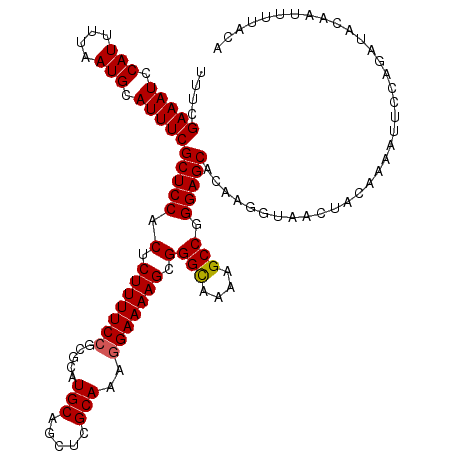
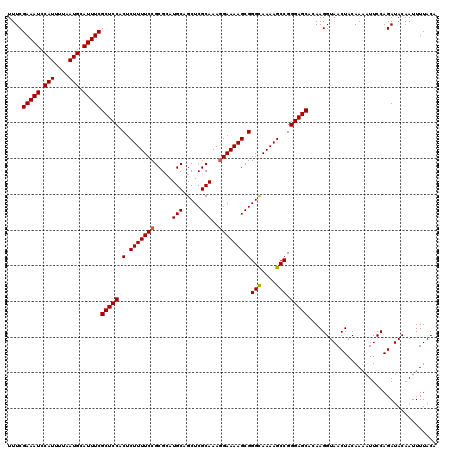
| Location | 2,463,496 – 2,463,610 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.27 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2463496 114 + 27905053 GAAAAGAGUGGAGCGAAAUGCAUUAAAAUGGAUUUCGAAACUUUAAAACUCAAUUUCAAUUGCACAGAAAAAGUUUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGAGCGGG (.(((..(..(((((((((.(((....))).)))))((((((((......((((....)))).......))))))))))))..)..))).).......(((........))).. ( -27.52) >DroSec_CAF1 22470 109 + 1 GAAAAGAGUGGAGCGAAAUGCAUUAAAAUGGAUUUCGAAACUUUAAAACUCAAUUUCAAUUGCACAGAAAAAGUUUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGA----- .......(..(((((((((.(((....))).)))))((((((((......((((....)))).......))))))))))))..)..((..(........)..)).....----- ( -26.22) >DroSim_CAF1 19210 109 + 1 GAAAAGUGUGGAGCGAAAUGCAUUAAAAUGGAUUUCGAAACUUUAAAACUCAAUUUCAAUUGCACAGAAAAAGUUUCGCUCUGCGAUUUGCAUAUUUACGUGGAAGGGA----- ......((..(((((((((.(((....))).)))))((((((((......((((....)))).......))))))))))))..)).((..(........)..)).....----- ( -26.62) >DroEre_CAF1 23914 109 + 1 GAAAAGAGUGGAGCGAAAUGCAUUAAAAUGGAUUUCGAAACUUUAAAACUCAAUUUCAAUUGCACAGAAAAAGUUUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGA----- .......(..(((((((((.(((....))).)))))((((((((......((((....)))).......))))))))))))..)..((..(........)..)).....----- ( -26.22) >consensus GAAAAGAGUGGAGCGAAAUGCAUUAAAAUGGAUUUCGAAACUUUAAAACUCAAUUUCAAUUGCACAGAAAAAGUUUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGA_____ .......(..(((((((((.(((....))).)))))((((((((......((((....)))).......))))))))))))..)..((..(........)..)).......... (-26.46 = -26.27 + -0.19)

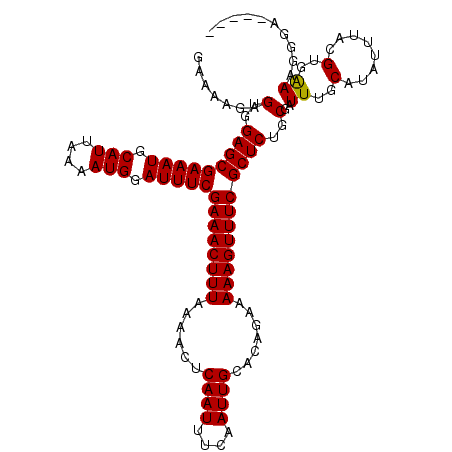
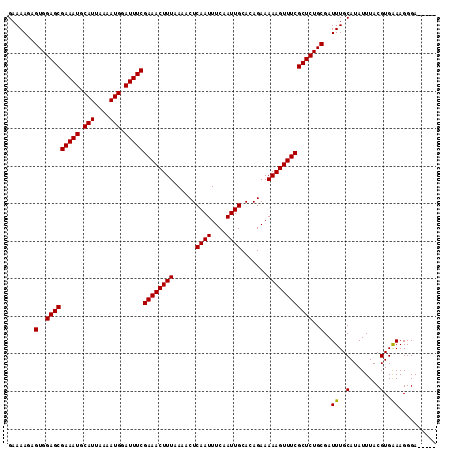
| Location | 2,463,496 – 2,463,610 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2463496 114 - 27905053 CCCGCUCCCUUUCACGUAAAUAUGCAAAUCGCAGAGCGAAACUUUUUCUGUGCAAUUGAAAUUGAGUUUUAAAGUUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUC ......................(((.....)))((((((((((((.......((((....))))......))))))))(((((.(((....))).))))))))).......... ( -21.32) >DroSec_CAF1 22470 109 - 1 -----UCCCUUUCACGUAAAUAUGCAAAUCGCAGAGCGAAACUUUUUCUGUGCAAUUGAAAUUGAGUUUUAAAGUUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUC -----.................(((.....)))((((((((((((.......((((....))))......))))))))(((((.(((....))).))))))))).......... ( -21.32) >DroSim_CAF1 19210 109 - 1 -----UCCCUUCCACGUAAAUAUGCAAAUCGCAGAGCGAAACUUUUUCUGUGCAAUUGAAAUUGAGUUUUAAAGUUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACACUUUUC -----..........((.....(((.....)))((((((((((((.......((((....))))......))))))))(((((.(((....))).))))))))).))....... ( -21.42) >DroEre_CAF1 23914 109 - 1 -----UCCCUUUCACGUAAAUAUGCAAAUCGCAGAGCGAAACUUUUUCUGUGCAAUUGAAAUUGAGUUUUAAAGUUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUC -----.................(((.....)))((((((((((((.......((((....))))......))))))))(((((.(((....))).))))))))).......... ( -21.32) >consensus _____UCCCUUUCACGUAAAUAUGCAAAUCGCAGAGCGAAACUUUUUCUGUGCAAUUGAAAUUGAGUUUUAAAGUUUCGAAAUCCAUUUUAAUGCAUUUCGCUCCACUCUUUUC ......................(((.....)))((((((((((((.......((((....))))......))))))))(((((.(((....))).))))))))).......... (-21.32 = -21.32 + 0.00)

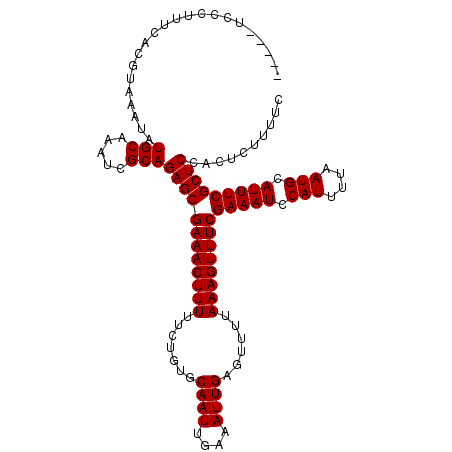
| Location | 2,463,570 – 2,463,690 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2463570 120 + 27905053 UUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGAGCGGGAGCUGAAAAACAAUAUCAUCCCGCGGCAGCACAAAAAAUGUAUGAAUUCCUAGGUCCACCUGGGAUGUUCAGAAAUUAAU ((((((((((..((..(........)..)).((((((....))(((.........))))))))))).)))............((((((((((((....))))))).))))))))...... ( -35.70) >DroSec_CAF1 22544 114 + 1 UUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGA------GCUGGAAAACAAUAUCAUCCCGCGGCAGCGCAAAAAAUGUAUGAAUUCCUAGGUCCACCUGGGAUGUUCAGAAAUUAAU ..((((((((..((..(........)..)).((((------....((........)).)))))))).)))).....(((...((((((((((((....))))))).)))))...)))... ( -34.20) >DroSim_CAF1 19284 114 + 1 UUCGCUCUGCGAUUUGCAUAUUUACGUGGAAGGGA------GCUGGAAAACAAUAUCAUCCCGCGGCAGCGCAAAAAAUGUAUAAAUUCCUAGGUCCACCUGGGAUGUUCAGAAAUUAAU ..((((((((..((..(........)..)).((((------....((........)).)))))))).))))................(((((((....)))))))............... ( -30.00) >DroEre_CAF1 23988 113 + 1 UUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGA------GCUGAAAAACAAAAUCAUCCCGCAGCAGCG-AAAAAACGUAUGAAUUCCUAGGUCCACCUGGGAUGUUCAGAAAUUAAU ((((((((((..((..(........)..)).((((------..(((.........))))))))))).))))-))........((((((((((((....))))))).)))))......... ( -36.60) >consensus UUCGCUCUGCGAUUUGCAUAUUUACGUGAAAGGGA______GCUGAAAAACAAUAUCAUCCCGCGGCAGCGCAAAAAAUGUAUGAAUUCCUAGGUCCACCUGGGAUGUUCAGAAAUUAAU ((((((((((..((..(........)..)).((((........((.....))......)))))))).)))............((((((((((((....))))))).))))))))...... (-31.26 = -31.14 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:27 2006