
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,187,321 – 18,187,481 |
| Length | 160 |
| Max. P | 0.696254 |

| Location | 18,187,321 – 18,187,441 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -31.84 |
| Energy contribution | -31.15 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18187321 120 - 27905053 AUUGAUCCACUGGGCACUAGUGCCUGGAACACGCUGAUCCGCGGCCACAAGCGCUGGUGCCUGUUCCCCACCCAAACGCCCAAGGAGCUGCUCAAGGUCACCAGUGCCAUGGGUGGCAAG .(((.......((((((((((((.(((....(((......))).)))...)))))))))))).....(((((((...(((....).)).......((.((....)))).)))))))))). ( -46.50) >DroVir_CAF1 589 120 - 1 AUUGAUCCAUUGGGUACCAGUGCCUGGAAUACGUUAAUACGUGGUCACAAGCGGUGGUGCUUAUUUCCCACGCACACGCCCAAGGACCUGCUGAAGGUCACCAGCGCUAUGGGCGGCAAG ..........(((((((((.(((.((((.(((((....))))).)).)).))).)))))))))........((...((((((.((....((((........)))).)).))))))))... ( -42.80) >DroGri_CAF1 620 120 - 1 AUAGAUCCGUUGGGUACCAGUGCCUGGAAUACGUUGAUACGCGGCCACAAACGCUGGUGCUUGUUUCCCACGCACACGCCUAAGGAUCUGUUGAAGGUGACAAGCGCAAUGGGCGGGAAG ((((((((.((((((..(((((..(((....(((......))).)))....)))))((((.((.....)).))))..))))))))))))))....(....)...(((.....)))..... ( -37.80) >DroWil_CAF1 12855 120 - 1 AUCGAUCCGCUGGGUACCAGCGCCUGGAAUACACUGAUACAUGGCCACAAACGUUGGUGCCUUUUUCCCACCCAGACGCCUAAGGAUCUUCUUAAGGUCACGAGCGCUAUGGGUGGCAAA ...........(((((((((((..(((....((........)).)))....))))))))))).....(((((((...((((...(((((.....)))))...)).))..))))))).... ( -40.70) >DroMoj_CAF1 593 120 - 1 AUAGAUCCGCUGGGCACCAGCGCAUGGAAUACCCUAGUUCGUGGCCACAAGCGCUGGUGCUUGUUUCCCACGCACACGCCCAAGGAUCUGCUCAAGGUUACUAGUGCUAUGGGAGGCAAG .(((((((..(((((.(((.....))).........((.(((((..(((((((....)))))))...))))).))..))))).)))))))((((.(((.......))).))))....... ( -46.00) >DroAna_CAF1 608 120 - 1 AUCGAUCCGCUAGGCACCAGUGCUUGGAACACUCUAAUCCGCGGUCACAAGCGGUGGUGCCUAUUCCCCACCCAAACACCGAAGGACUUGCUGAAAGUGACCAGUGCCAUGGGUGGAAAG ..........(((((((((.((((((..((.(.(......).)))..)))))).)))))))))....(((((((..((((....)((((.....)))).....)))...))))))).... ( -45.20) >consensus AUAGAUCCGCUGGGCACCAGUGCCUGGAAUACGCUAAUACGCGGCCACAAGCGCUGGUGCCUGUUUCCCACCCACACGCCCAAGGAUCUGCUGAAGGUCACCAGCGCUAUGGGUGGCAAG ......((..(((((((((((((.((.....(((......)))....)).)))))))))))))...((((......(((.....(((((.....)))))....)))...)))).)).... (-31.84 = -31.15 + -0.69)


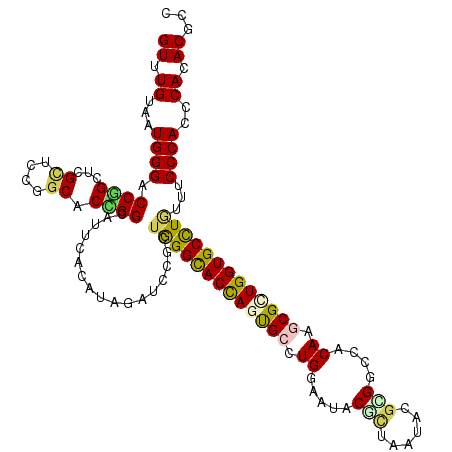
| Location | 18,187,361 – 18,187,481 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -31.63 |
| Energy contribution | -31.13 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18187361 120 - 27905053 GUUUGUCAUGGGACCGGCUCGCUCCGGCACCGGCAUCCACAUUGAUCCACUGGGCACUAGUGCCUGGAACACGCUGAUCCGCGGCCACAAGCGCUGGUGCCUGUUCCCCACCCAAACGCC (((((...((((.((((......))))....((.(((......)))))...((((((((((((.(((....(((......))).)))...))))))))))))....))))..)))))... ( -46.40) >DroVir_CAF1 629 120 - 1 GUUUGUAAUGGGUCCAGCUCGCUCCGGCACUGGUAUUCAUAUUGAUCCAUUGGGUACCAGUGCCUGGAAUACGUUAAUACGUGGUCACAAGCGGUGGUGCUUAUUUCCCACGCACACGCC ((((((...(((.....)))..(((((((((((((((((...........)))))))))))))).))).(((((....)))))...))))))((((((((...........)))).)))) ( -47.20) >DroGri_CAF1 660 120 - 1 GUUUGUAAUGGGCCCGGCACGUUCUGGCACCGGUAUUCACAUAGAUCCGUUGGGUACCAGUGCCUGGAAUACGUUGAUACGCGGCCACAAACGCUGGUGCUUGUUUCCCACGCACACGCC ((((((....((((((.(((((((((((((.(((((((((........).)))))))).))))).)))..))).))...)).))))))))))((..((((.((.....)).))))..)). ( -45.20) >DroWil_CAF1 12895 120 - 1 GUUUGUUAUGGGACCCGGACGCUCUGGCACGGGCAUACAUAUCGAUCCGCUGGGUACCAGCGCCUGGAAUACACUGAUACAUGGCCACAAACGUUGGUGCCUUUUUCCCACCCAGACGCC (((((...(((((((((...((....)).))))..................(((((((((((..(((....((........)).)))....)))))))))))...)))))..)))))... ( -38.50) >DroMoj_CAF1 633 120 - 1 GUUUGUCAUGGGCCCGGCUCGUUCCGGCACCGGCAUUCAUAUAGAUCCGCUGGGCACCAGCGCAUGGAAUACCCUAGUUCGUGGCCACAAGCGCUGGUGCUUGUUUCCCACGCACACGCC ((.((((.((((.((((......))))..(((((..((.....))...)))))((((((((((...((((......))))((....))..))))))))))......)))).).))).)). ( -44.60) >DroAna_CAF1 648 120 - 1 GUUUGUUAUGGGGCCUGCUCGGUCCGGCACCGGUAUCCACAUCGAUCCGCUAGGCACCAGUGCUUGGAACACUCUAAUCCGCGGUCACAAGCGGUGGUGCCUAUUCCCCACCCAAACACC (((((...(((((.......((.(((....)))...))............(((((((((.((((((..((.(.(......).)))..)))))).)))))))))..)))))..)))))... ( -48.00) >consensus GUUUGUAAUGGGACCGGCUCGCUCCGGCACCGGCAUUCACAUAGAUCCGCUGGGCACCAGUGCCUGGAAUACGCUAAUACGCGGCCACAAGCGCUGGUGCCUGUUUCCCACCCACACGCC ((.((...((((.((((...((....)).)))).................(((((((((((((.((.....(((......)))....)).)))))))))))))...))))..)).))... (-31.63 = -31.13 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:17 2006