
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,184,113 – 18,184,256 |
| Length | 143 |
| Max. P | 0.858460 |

| Location | 18,184,113 – 18,184,225 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -25.74 |
| Energy contribution | -26.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18184113 112 - 27905053 AA--------GGAAACUCACUUGGAUGCAGUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAGAGUGUGGUUUUGCCAGCAUUAUCCAGGCCCAGGAACAAUAAUUU .(--------(....))..(((((..((((((((....))))))))(((.(((((.......))))).........(((((((.....))).)))).....))))))))........... ( -35.20) >DroPse_CAF1 2884 113 - 1 GA-------CUGCAACUCACUUGGAUGCAGGGUGGGGACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUGUGCAGUAGCGUGGUCUUGCCAGCAUUAUCCAAGCCAAGGAACAACAAUUU ..-------..........(((((((((..((..((..(((((((((((((((((.......))))).)).)).))))))))..))..)).)))...))))))................. ( -41.00) >DroGri_CAF1 337 120 - 1 UCUGAAUCGCUUUCACUCACUUGGAUGCAGUGUUGGCACAUGCUGCGCUAGCUUAUCAUCUUUGAGCAUAUGCAAUAAUGUGGUUUUACCAGCAUUAUCCAAGCCAAGGAACAAUAGUUU ((((((.....))).....((((((((..((((((((((((..((((...(((((.......)))))...))))...)))))......)))))))))))))))....))).......... ( -30.90) >DroWil_CAF1 720 113 - 1 AA-------UUUCUACUUACUUGGAUGCAAUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAUAAUGUGGUUUUACCAGCAUUAUCCAAGCCAAGGAACAAUAAUUU ..-------......(((.((((((((((...((((((.....))).)))(((((.......)))))...)))).((((((((.....))).)))))))))))..)))............ ( -29.40) >DroAna_CAF1 541 111 - 1 -U--------GAAUACUUACUCGGAUGCAGGGUGGGCACAUGCUGGGCCAGCUUGUCAUCUUUGAGCAUGUGCAACAGUGUGGUUUUUCCAGCAUUGUCCAAGCCGAGGAACAACAACUU -.--------.........(((((........(((((((((((.(((....))).(((....))))))))))).(((((((((.....))).)))))))))..)))))............ ( -33.40) >DroPer_CAF1 3618 113 - 1 GA-------CUGCAACUCACUUGGAUGCAGGGUGGGGACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUGUGCAGUAGCGUGGUCUUGCCAGCAUUAUCCAAGCCAAGGAACAACAAUUU ..-------..........(((((((((..((..((..(((((((((((((((((.......))))).)).)).))))))))..))..)).)))...))))))................. ( -41.00) >consensus AA_______CUGCAACUCACUUGGAUGCAGGGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAUAGUGUGGUUUUGCCAGCAUUAUCCAAGCCAAGGAACAACAAUUU ...................(((((..((((.(((....))).))))(((((((((.......))))).)).)).(((((((((.....))).)))))).....)))))............ (-25.74 = -26.05 + 0.31)

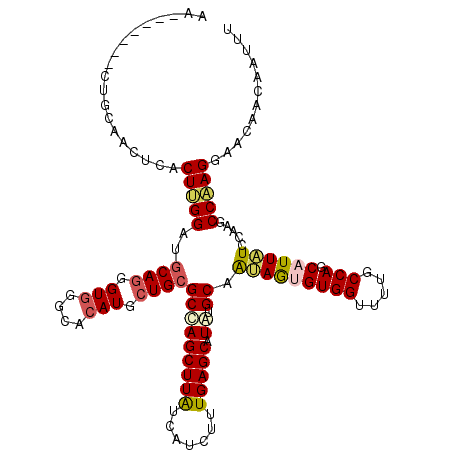

| Location | 18,184,153 – 18,184,256 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -20.03 |
| Energy contribution | -21.03 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18184153 103 - 27905053 AGUCACACACAUAACCACUAUUUAGUCCAUG-------AAGGAAACUCACUUGGAUGCAGUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAGAGUG .......(((....((((......(((((((-------(.(....))))..))))).....))))(((.((((.((....))...(((....))))))).)))....))) ( -28.20) >DroVir_CAF1 533 95 - 1 UA----AUACAUAA------UAUAGUAGCUG-----CAUUUAAGACUUACUUGGAUGCAGUGUUGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAUAAUG ..----...((((.------....(((((((-----((((((((.....))))))))))(((....)))..)))))....(((((.......))))).))))........ ( -27.00) >DroSec_CAF1 917 99 - 1 GU----AUGCACAAUGAGGAUUUUGUCCAGG-------GAGCAGACUCACUUGGAUGCAGUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAGAGUG ((----((((.(((.((.(((...(((((((-------(((....))).)))))))((((((((....))))))))........))).)).))).))))))......... ( -34.60) >DroSim_CAF1 802 99 - 1 GU----AUACACAUUGAGGAUUUUGUCCAGG-------GAGCAGACUCACUUGGAUGCAGUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAGAGUG ((----(((........((.....(((((((-------(((....))).)))))))((((((((....)))))))).)).(((((.......))))).)))))....... ( -33.70) >DroYak_CAF1 989 104 - 1 AG----UUACACAACGA--AUUCCGUCCAGGAAGGGAGGAGCAGACUCACUUGGAUGCAGUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAGAGUG ..----...(((.....--..(((..((.....))..)))(((........(((.(((((((((....))))))))))))(((((.......)))))...)))....))) ( -33.20) >DroMoj_CAF1 433 100 - 1 AG----CUACAGAACAA-UCUGGAGUAGAUG-----AGUUUUACACUUACUUGGAUGCAGUGUUGGGACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAUAAUG .(----((.((((....-)))).))).((((-----(((............(((.((((((((......))))))))))))))))))........((....))....... ( -24.40) >consensus AG____AUACACAACGA__AUUUAGUCCAGG_______GAGCAGACUCACUUGGAUGCAGUGUGGGCACAUGCUGCGCCAGCUUAUCAUCUUUGAGCAUAUGCAAGAGUG ............................................((((...(((.(((((((((....))))))))))))(((((.......)))))........)))). (-20.03 = -21.03 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:14 2006