
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,180,046 – 18,180,158 |
| Length | 112 |
| Max. P | 0.824210 |

| Location | 18,180,046 – 18,180,158 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.24 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
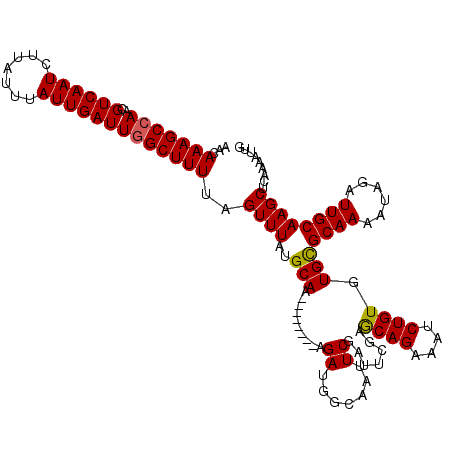
>3R_DroMel_CAF1 18180046 112 + 27905053 AACAAAGCCAACGUCAAUCUUAUUUAUUGAUUGGCUUUUAGUUUACGCAA--------AGAUGGCAAUUCGAUUCGAACAGAAAUCUGUGUGCGCAAAAUAGAUUGCAAGCUCAAAAUUG ...(((((((..((((((.......))))))))))))).(((((..(((.--------((((..(..((((...))))..)..))))...)))((((......)))))))))........ ( -25.60) >DroSec_CAF1 2917 112 + 1 AACAAAGCCAACGUCAAUCUUAUUUAUUGAUUGGCUUUUAGUUUAUGCAA--------AGAUGGCAAUUCGAUUCGAGCAGAAAUCUGUGUGUGCAAAGUAAAUUGCAAGCUCAAAAUUG ......((((.((((((((.........))))))).....((....))..--------.).))))..........((((((....)).....(((((......))))).))))....... ( -24.30) >DroSim_CAF1 2000 112 + 1 AAAAAAGCAAACGUCAAUCUUAUUUAUUGAUUGGCUUUUAGUUUAUGCAA--------AGAUGGCAAUUCGAUUCGAACAGAAAUCUGUGUGCGCAAAAUAGAUUGCAAGCUCAAAAUUG ......((((..(((((((.........))))))).((((.(((.((((.--------((((..(..((((...))))..)..))))...)))).))).))))))))............. ( -22.90) >DroEre_CAF1 1974 112 + 1 AACAAAGCCAACGUCAAUCUUAUUUAUUGAUUGGCUUUUAGUUUAUGCAA--------AGAUCGCAAUUCGAUUCGAGCAGAAAUCUGUGUGCGCAAAAUAGAUUGCAAGCACAAAAUGG ...(((((((..((((((.......)))))))))))))((((((.(((..--------.(((((.....)))))...))).))).)))(((((((((......))))..)))))...... ( -26.40) >DroYak_CAF1 2058 120 + 1 AACAAAGCCAACGUCAAUCUUAUUUAUUGAUUGGCUUUCAGUUUAUGCAAAGAUGCAAAGAUGGCAAUUCGAUUCGUGCAGAAAUCUGUGUGCGCAAAAUAGAUUGCAAGCGCCAAAUGG ...(((((((..((((((.......)))))))))))))..((((.((((....)))).))))(((.........((..(((....)))..)).((((......))))....)))...... ( -30.40) >consensus AACAAAGCCAACGUCAAUCUUAUUUAUUGAUUGGCUUUUAGUUUAUGCAA________AGAUGGCAAUUCGAUUCGAGCAGAAAUCUGUGUGCGCAAAAUAGAUUGCAAGCUCAAAAUUG ...(((((((..((((((.......)))))))))))))..((((..(((..........((.......)).......((((....)))).)))((((......))))))))......... (-20.44 = -20.24 + -0.20)


| Location | 18,180,046 – 18,180,158 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
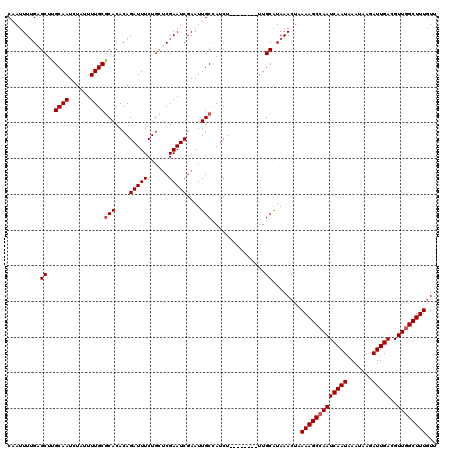
>3R_DroMel_CAF1 18180046 112 - 27905053 CAAUUUUGAGCUUGCAAUCUAUUUUGCGCACACAGAUUUCUGUUCGAAUCGAAUUGCCAUCU--------UUGCGUAAACUAAAAGCCAAUCAAUAAAUAAGAUUGACGUUGGCUUUGUU ......................((((((((...((((..(.(((((...))))).)..))))--------.))))))))...(((((((((((((.......)))))..))))))))... ( -24.80) >DroSec_CAF1 2917 112 - 1 CAAUUUUGAGCUUGCAAUUUACUUUGCACACACAGAUUUCUGCUCGAAUCGAAUUGCCAUCU--------UUGCAUAAACUAAAAGCCAAUCAAUAAAUAAGAUUGACGUUGGCUUUGUU ....(((((((.(((((......))))).....((....)))))))))......(((.....--------..))).......(((((((((((((.......)))))..))))))))... ( -24.10) >DroSim_CAF1 2000 112 - 1 CAAUUUUGAGCUUGCAAUCUAUUUUGCGCACACAGAUUUCUGUUCGAAUCGAAUUGCCAUCU--------UUGCAUAAACUAAAAGCCAAUCAAUAAAUAAGAUUGACGUUUGCUUUUUU .....(((.((((((((......))))(((...((((..(.(((((...))))).)..))))--------.))).........)))))))(((((.......)))))............. ( -17.80) >DroEre_CAF1 1974 112 - 1 CCAUUUUGUGCUUGCAAUCUAUUUUGCGCACACAGAUUUCUGCUCGAAUCGAAUUGCGAUCU--------UUGCAUAAACUAAAAGCCAAUCAAUAAAUAAGAUUGACGUUGGCUUUGUU ....(((((((..((((......))))(((....(((((......)))))....))).....--------..)))))))...(((((((((((((.......)))))..))))))))... ( -27.80) >DroYak_CAF1 2058 120 - 1 CCAUUUGGCGCUUGCAAUCUAUUUUGCGCACACAGAUUUCUGCACGAAUCGAAUUGCCAUCUUUGCAUCUUUGCAUAAACUGAAAGCCAAUCAAUAAAUAAGAUUGACGUUGGCUUUGUU .....((((((..((((......)))))).....(((((......))))).....))))....((((....)))).......(((((((((((((.......)))))..))))))))... ( -28.20) >consensus CAAUUUUGAGCUUGCAAUCUAUUUUGCGCACACAGAUUUCUGCUCGAAUCGAAUUGCCAUCU________UUGCAUAAACUAAAAGCCAAUCAAUAAAUAAGAUUGACGUUGGCUUUGUU .........((..((((......))))(((....(((((......)))))....)))...............))........(((((((((((((.......)))))..))))))))... (-19.88 = -20.28 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:10 2006