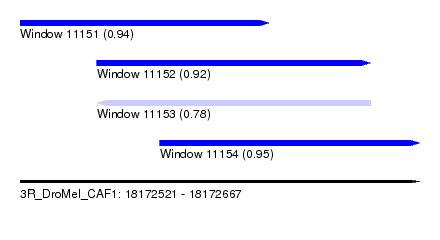
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,172,521 – 18,172,667 |
| Length | 146 |
| Max. P | 0.952162 |

| Location | 18,172,521 – 18,172,612 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -15.95 |
| Energy contribution | -17.05 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18172521 91 + 27905053 CCAAAAAGCAAAAGUUU------GC------ACCGCCAGUGC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAA .......((((....))------))------...((((.(((-----------------(((((...))))..((((((....))))))))))..)))).(((((((.....))))))). ( -25.70) >DroGri_CAF1 217034 114 + 1 CCAAAAAGCAAAAGUUU------GCCUUCAGGCUGCAAGUGGACGUCGAGACGCAUUUGUAUACACAAUGUUUGUGCUGAUUGCAGCGCAGCAAAUAGCUUCUGAUUUCACAAAUCAAAA .............((..------(((....))).))..(((((.(((.(((.((((((((..((.....)).(((((((....))))))))))))).)).)))))))))))......... ( -29.60) >DroEre_CAF1 216676 91 + 1 CCAAAAAGCAAAAGUUU------GC------ACCGCCAGUGC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAA .......((((....))------))------...((((.(((-----------------(((((...))))..((((((....))))))))))..)))).(((((((.....))))))). ( -25.70) >DroYak_CAF1 211228 91 + 1 CCAAAAAGCAAAAGUUU------GC------ACCGCCAGUGC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAA .......((((....))------))------...((((.(((-----------------(((((...))))..((((((....))))))))))..)))).(((((((.....))))))). ( -25.70) >DroMoj_CAF1 257710 100 + 1 CCAAAAAGCAAAAGUUUUUUGCUGCCUGCUGCCUGC----UGACUGC----------------UACAAUGUUUGUGCUGAUUGCAGCGCUGCAAAUAGCUUUUGAUUUCACAAAUCAAAA ..(((((((....)))))))((((..(((.((((((----.....((----------------.(((.....))))).....)))).)).)))..)))).(((((((.....))))))). ( -27.10) >DroAna_CAF1 195853 91 + 1 CCAAAAAGCAAAAGUUU------CC------ACUGCCAGUGC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAA ........((((((((.------..------..(((..((((-----------------((((((((.....)))).....)))))))).)))...))))))))................ ( -23.00) >consensus CCAAAAAGCAAAAGUUU______GC______ACCGCCAGUGC_________________UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAA ..................................((((((((..................))))....((((.((((((....))))))))))..)))).(((((((.....))))))). (-15.95 = -17.05 + 1.10)



| Location | 18,172,549 – 18,172,649 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18172549 100 + 27905053 GC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCAG ((-----------------.(((((...((((.((((((....))))))))))...(((((((((((.....)))))))))))((.((((((---...)))))).))..)))))..)).. ( -34.80) >DroGri_CAF1 217068 115 + 1 GGACGUCGAGACGCAUUUGUAUACACAAUGUUUGUGCUGAUUGCAGCGCAGCAAAUAGCUUCUGAUUUCACAAAUCAAAAGUUUCUUGUGC-----GCCGCACGUGAUUUAUGCCUGCAG ((.((((((((.((..((((....))))....(((((((....)))))))))....(((((.(((((.....))))).)))))))))).))-----)))(((.((.......)).))).. ( -31.80) >DroEre_CAF1 216704 100 + 1 GC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCGG ((-----------------.(((((...((((.((((((....))))))))))...(((((((((((.....)))))))))))...))))))---).(((((.((.......)).))))) ( -35.30) >DroYak_CAF1 211256 100 + 1 GC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCGG ((-----------------.(((((...((((.((((((....))))))))))...(((((((((((.....)))))))))))...))))))---).(((((.((.......)).))))) ( -35.30) >DroMoj_CAF1 257746 99 + 1 UGACUGC----------------UACAAUGUUUGUGCUGAUUGCAGCGCUGCAAAUAGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGC-----AGCGCACGUGAUUUAUGCCUGCAG ...((((----------------.(((.....)))((.(((..(.((((((((...(((((((((((.....))))))))))).....)))-----)))))..)..)))...))..)))) ( -35.50) >DroAna_CAF1 195881 103 + 1 GC-----------------UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCGCCGCGCCGCACGUGAUUUAUGCCUGCGG ((-----------------.((.(((((((((.((((((....))))))))))...(((((((((((.....)))))))))))..))))).)).)).(((((.((.......)).))))) ( -36.90) >consensus GC_________________UGCAUACAAUGUUUGUGCUGAUUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG___CGCCGCACGUGAUUUAUGCCUGCAG ................................(((((((....)))))))(((...(((((((((((.....)))))))))))((.((((((......)))))).))........))).. (-28.04 = -28.27 + 0.22)



| Location | 18,172,549 – 18,172,649 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18172549 100 - 27905053 CUGCAGGCAUAAAUCACGUGCGGCG---CGCACAAGAAACUUUUGAUUUGUGAAAUCAAAAGCCAUUUGCUGUGCUGCAAUCAGCACAAACAUUGUAUGCA-----------------GC ((((((((.........(((((...---))))).......((((((((.....)))))))))))......(((((((....))))))).........))))-----------------). ( -32.00) >DroGri_CAF1 217068 115 - 1 CUGCAGGCAUAAAUCACGUGCGGC-----GCACAAGAAACUUUUGAUUUGUGAAAUCAGAAGCUAUUUGCUGCGCUGCAAUCAGCACAAACAUUGUGUAUACAAAUGCGUCUCGACGUCC ......((((........((((((-----(((((((...(((((((((.....)))))))))...)))).)))))))))....(((((.....)))))......))))............ ( -34.50) >DroEre_CAF1 216704 100 - 1 CCGCAGGCAUAAAUCACGUGCGGCG---CGCACAAGAAACUUUUGAUUUGUGAAAUCAAAAGCCAUUUGCUGUGCUGCAAUCAGCACAAACAUUGUAUGCA-----------------GC (((((.(.........).))))).(---((.((((....(((((((((.....)))))))))........(((((((....)))))))....)))).))).-----------------.. ( -29.80) >DroYak_CAF1 211256 100 - 1 CCGCAGGCAUAAAUCACGUGCGGCG---CGCACAAGAAACUUUUGAUUUGUGAAAUCAAAAGCCAUUUGCUGUGCUGCAAUCAGCACAAACAUUGUAUGCA-----------------GC (((((.(.........).))))).(---((.((((....(((((((((.....)))))))))........(((((((....)))))))....)))).))).-----------------.. ( -29.80) >DroMoj_CAF1 257746 99 - 1 CUGCAGGCAUAAAUCACGUGCGCU-----GCACAAGAAACUUUUGAUUUGUGAAAUCAAAAGCUAUUUGCAGCGCUGCAAUCAGCACAAACAUUGUA----------------GCAGUCA .(((((((((.......))))(((-----(((.((....(((((((((.....)))))))))...)))))))).)))))....(((((.....))).----------------))..... ( -29.40) >DroAna_CAF1 195881 103 - 1 CCGCAGGCAUAAAUCACGUGCGGCGCGGCGCACAAGAAACUUUUGAUUUGUGAAAUCAAAAGCCAUUUGCUGUGCUGCAAUCAGCACAAACAUUGUAUGCA-----------------GC ..(((.(((........((((...((((((((((((...(((((((((.....)))))))))...)))).)))))))).....))))......))).))).-----------------.. ( -33.64) >consensus CCGCAGGCAUAAAUCACGUGCGGCG___CGCACAAGAAACUUUUGAUUUGUGAAAUCAAAAGCCAUUUGCUGUGCUGCAAUCAGCACAAACAUUGUAUGCA_________________GC ..((((((.........(((((......))))).......((((((((.....)))))))))))...)))(((((((....)))))))................................ (-24.36 = -24.83 + 0.47)


| Location | 18,172,572 – 18,172,667 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -26.65 |
| Energy contribution | -26.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18172572 95 + 27905053 UUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCAGCAGGAGC-AAGUCCAGAU---U ((((.((((((.....(((((((((((.....)))))))))))..)))))).---.((.(((.((.......)).))).))....))-))........---. ( -27.60) >DroSim_CAF1 208673 95 + 1 UUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCGGCAGGAGC-AAGUCCAGAU---U ((((.((((((.....(((((((((((.....)))))))))))..)))))).---.((((((.((.......)).))))))....))-))........---. ( -33.70) >DroEre_CAF1 216727 95 + 1 UUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCGGCAGGGGC-AAGUCCAGAU---U ..((.((((((.....(((((((((((.....)))))))))))..)))))))---)((((((.((.......)).))))))..(((.-...)))....---. ( -33.90) >DroYak_CAF1 211279 95 + 1 UUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG---CGCCGCACGUGAUUUAUGCCUGCGGCAGGAGC-AAGUCCAGAU---U ((((.((((((.....(((((((((((.....)))))))))))..)))))).---.((((((.((.......)).))))))....))-))........---. ( -33.70) >DroMoj_CAF1 257770 89 + 1 UUGCAGCGCUGCAAAUAGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGC-----AGCGCACGUGAUUUAUGCCUGCAGCAG--------UCCAAAUGUCU .....((((((((...(((((((((((.....))))))))))).....)))-----)))))((((......(((.....))).--------.....)))).. ( -30.90) >DroAna_CAF1 195904 99 + 1 UUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCGCCGCGCCGCACGUGAUUUAUGCCUGCGGCAGGAGCCGAGUCCAGAU---U ..((.((.((.(((..(((((((((((.....)))))))))))..))))).)).))((((((.((.......)).)))))).(((......)))....---. ( -34.40) >consensus UUGCAGCACAGCAAAUGGCUUUUGAUUUCACAAAUCAAAAGUUUCUUGUGCG___CGCCGCACGUGAUUUAUGCCUGCGGCAGGAGC_AAGUCCAGAU___U .(((.(((..(((...(((((((((((.....)))))))))))((.((((((......)))))).))....))).))).)))(((......)))........ (-26.65 = -26.77 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:03 2006