
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,169,888 – 18,170,082 |
| Length | 194 |
| Max. P | 0.995273 |

| Location | 18,169,888 – 18,169,988 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18169888 100 + 27905053 UGUGUGAAGUGCGGCUCGUUGGCGCUUUUUAGUAAAAUUACACGCUCUAAUCCACAACUUUUGAGUGGGCCCACACCACCGAAAAGUGUGGAAAAAAA--------AG .(((((((((((.(.....).)))))))...(((....))))))).....((((((..(((((.((((.......)))))))))..))))))......--------.. ( -29.70) >DroSec_CAF1 198119 97 + 1 UGUGUGAAGUGCGGCUCGUUGGCGCUUUUUAGUAAAAUUACACGCUCUAAUCCACAACUUUUGAGUGGGCCCACACCACCGAAAAGUGUUGAAA-A----------AA .(((((((((((.(.....).)))))))...(((....))))))).....((.(((..(((((.((((.......)))))))))..))).))..-.----------.. ( -23.20) >DroSim_CAF1 205964 98 + 1 UGUGUGAAGUGCGGCUCGUUGGCGCUUUUUAGUAAAAUUACACGCUCUAAUCCACAACUUUUGAGUGGGCCCACACCACCGAAAAGUGUGGAAAAA----------AA .(((((((((((.(.....).)))))))...(((....))))))).....((((((..(((((.((((.......)))))))))..))))))....----------.. ( -29.70) >DroEre_CAF1 213978 98 + 1 UGUGUGAAGUGCGGCUCGUUGGCGCUUUUUAGUGAAAUUACACGCUCUAAUCCACAACUUUUGAGUGGGCCCACACCAUCGAAAAGUGUGGAAAAA----------AG .((((((...(((.(.....).)))((((....)))))))))).......((((((.(((((..((((.......))))..)))))))))))....----------.. ( -28.00) >DroYak_CAF1 208474 108 + 1 UGUGUGAAGUGCGGCUCGCUGGCGCUUUUUAGUAAAAUUACACGCUCUAAUCCACAACUUUUGAGUGGGCCCACACCACCGAAAAGUGUGGAAAAAAAAAGAAAAGAG .(((((((((((((....)).)))))))...(((....))))))).....((((((..(((((.((((.......)))))))))..))))))................ ( -30.60) >consensus UGUGUGAAGUGCGGCUCGUUGGCGCUUUUUAGUAAAAUUACACGCUCUAAUCCACAACUUUUGAGUGGGCCCACACCACCGAAAAGUGUGGAAAAA__________AG .(((((((((((.(.....).)))))))...(((....))))))).....((((((..(((((.((((.......)))))))))..))))))................ (-28.16 = -28.04 + -0.12)



| Location | 18,169,888 – 18,169,988 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18169888 100 - 27905053 CU--------UUUUUUUCCACACUUUUCGGUGGUGUGGGCCCACUCAAAAGUUGUGGAUUAGAGCGUGUAAUUUUACUAAAAAGCGCCAACGAGCCGCACUUCACACA ..--------((((..(((((((((((.(((((.(....)))))).))))).))))))..)))).((((..((((....))))(((.(.....).))).....)))). ( -27.10) >DroSec_CAF1 198119 97 - 1 UU----------U-UUUCAACACUUUUCGGUGGUGUGGGCCCACUCAAAAGUUGUGGAUUAGAGCGUGUAAUUUUACUAAAAAGCGCCAACGAGCCGCACUUCACACA ..----------.-...............((((.((((((((((.........))))....(.((((................))))).....))).))).))))... ( -21.39) >DroSim_CAF1 205964 98 - 1 UU----------UUUUUCCACACUUUUCGGUGGUGUGGGCCCACUCAAAAGUUGUGGAUUAGAGCGUGUAAUUUUACUAAAAAGCGCCAACGAGCCGCACUUCACACA ((----------((..(((((((((((.(((((.(....)))))).))))).))))))..)))).((((..((((....))))(((.(.....).))).....)))). ( -26.90) >DroEre_CAF1 213978 98 - 1 CU----------UUUUUCCACACUUUUCGAUGGUGUGGGCCCACUCAAAAGUUGUGGAUUAGAGCGUGUAAUUUCACUAAAAAGCGCCAACGAGCCGCACUUCACACA ((----------((..(((((((((((.(((((.(....)))).))))))).))))))..)))).(((......)))......(((.(.....).))).......... ( -23.50) >DroYak_CAF1 208474 108 - 1 CUCUUUUCUUUUUUUUUCCACACUUUUCGGUGGUGUGGGCCCACUCAAAAGUUGUGGAUUAGAGCGUGUAAUUUUACUAAAAAGCGCCAGCGAGCCGCACUUCACACA ..........((((..(((((((((((.(((((.(....)))))).))))).))))))..)))).((((..............((....))(((......))))))). ( -27.40) >consensus CU__________UUUUUCCACACUUUUCGGUGGUGUGGGCCCACUCAAAAGUUGUGGAUUAGAGCGUGUAAUUUUACUAAAAAGCGCCAACGAGCCGCACUUCACACA ................(((((((((((.(((((.(....)))))).))))).)))))).......((((..((((....))))(((.(.....).))).....)))). (-22.68 = -23.28 + 0.60)



| Location | 18,169,924 – 18,170,019 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -14.73 |
| Energy contribution | -16.53 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18169924 95 + 27905053 AUUACACGCUCUAAUCCACAACUUU-UGAGUGGGC---CCACACCACCGAAAAGUGUGGAAAAAAA--------AGGGGCCAGAAUCCAGAA-AACCCUCUGACGAGU .......((((...((((((..(((-((.((((..---.....)))))))))..))))))......--------((((..............-..)))).....)))) ( -27.09) >DroSec_CAF1 198155 92 + 1 AUUACACGCUCUAAUCCACAACUUU-UGAGUGGGC---CCACACCACCGAAAAGUGUUGAAA-A----------AAGGGCCAGAAUCUAGAA-AACCCUCUGACGAGU .......(((((..((.(((..(((-((.((((..---.....)))))))))..))).))..-.----------.)))))((((........-.....))))...... ( -19.82) >DroSim_CAF1 206000 93 + 1 AUUACACGCUCUAAUCCACAACUUU-UGAGUGGGC---CCACACCACCGAAAAGUGUGGAAAAA----------AAGGGCCAGAAUCUAGAA-AACCCUCUGACGAGU .......((((...((((((..(((-((.((((..---.....)))))))))..))))))....----------.((((.............-..)))).....)))) ( -25.86) >DroEre_CAF1 214014 93 + 1 AUUACACGCUCUAAUCCACAACUUU-UGAGUGGGC---CCACACCAUCGAAAAGUGUGGAAAAA----------AGCGGCCAGAAUCCAGAA-AACCCUCUGACGAGU ......((((....((((((.((((-(..((((..---.....))))..)))))))))))....----------)))).........((((.-.....))))...... ( -26.00) >DroYak_CAF1 208510 104 + 1 AUUACACGCUCUAAUCCACAACUUU-UGAGUGGGC---CCACACCACCGAAAAGUGUGGAAAAAAAAAGAAAAGAGGGGCCAGAAUCCAGAAAAACCCUCUGACGAGU .......((((...((((((..(((-((.((((..---.....)))))))))..))))))............((((((.................))))))...)))) ( -32.13) >DroMoj_CAF1 254709 91 + 1 AUUACACGCUCUAAUCCACAAGUUUGCGAGUGGAGAGCCCGUCCAACC--------UGGCAGACAAAAUGAA-------AGAGAACCUAA-A-AACAAACCAAAGAGA ........((((.........((((((.((((((.......))))..)--------).))))))........-------((.....))..-.-..........)))). ( -15.70) >consensus AUUACACGCUCUAAUCCACAACUUU_UGAGUGGGC___CCACACCACCGAAAAGUGUGGAAAAA__________AGGGGCCAGAAUCCAGAA_AACCCUCUGACGAGU .......((((...((((((.((((.((.((((..........)))))).))))))))))...........................((((.......))))..)))) (-14.73 = -16.53 + 1.81)
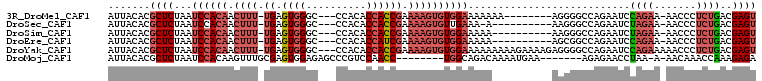


| Location | 18,169,924 – 18,170,019 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -17.65 |
| Energy contribution | -19.65 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18169924 95 - 27905053 ACUCGUCAGAGGGUU-UUCUGGAUUCUGGCCCCU--------UUUUUUUCCACACUUUUCGGUGGUGUGG---GCCCACUCA-AAAGUUGUGGAUUAGAGCGUGUAAU ((.(((.((.(((((-...........)))))))--------.(((..(((((((((((.(((((.(...---.)))))).)-)))).))))))..)))))).))... ( -29.20) >DroSec_CAF1 198155 92 - 1 ACUCGUCAGAGGGUU-UUCUAGAUUCUGGCCCUU----------U-UUUCAACACUUUUCGGUGGUGUGG---GCCCACUCA-AAAGUUGUGGAUUAGAGCGUGUAAU ((.((((((((((((-...........)))))))----------)-.((((..((((((.(((((.(...---.)))))).)-)))))..))))...)).)).))... ( -24.70) >DroSim_CAF1 206000 93 - 1 ACUCGUCAGAGGGUU-UUCUAGAUUCUGGCCCUU----------UUUUUCCACACUUUUCGGUGGUGUGG---GCCCACUCA-AAAGUUGUGGAUUAGAGCGUGUAAU ((.((((((((((((-...........)))))))----------)...(((((((((((.(((((.(...---.)))))).)-)))).))))))...)).)).))... ( -30.60) >DroEre_CAF1 214014 93 - 1 ACUCGUCAGAGGGUU-UUCUGGAUUCUGGCCGCU----------UUUUUCCACACUUUUCGAUGGUGUGG---GCCCACUCA-AAAGUUGUGGAUUAGAGCGUGUAAU ....(((((((..(.-....)..)))))))((((----------((..(((((((((((.(((((.(...---.)))).)))-)))).))))))..))))))...... ( -31.40) >DroYak_CAF1 208510 104 - 1 ACUCGUCAGAGGGUUUUUCUGGAUUCUGGCCCCUCUUUUCUUUUUUUUUCCACACUUUUCGGUGGUGUGG---GCCCACUCA-AAAGUUGUGGAUUAGAGCGUGUAAU ((.(((.((((((.....(..(...)..).)))))).......(((..(((((((((((.(((((.(...---.)))))).)-)))).))))))..)))))).))... ( -31.50) >DroMoj_CAF1 254709 91 - 1 UCUCUUUGGUUUGUU-U-UUAGGUUCUCU-------UUCAUUUUGUCUGCCA--------GGUUGGACGGGCUCUCCACUCGCAAACUUGUGGAUUAGAGCGUGUAAU ...............-.-.((.(((((..-------(((((...((.(((.(--------(..((((.......)))))).))).))..)))))..))))).)).... ( -19.00) >consensus ACUCGUCAGAGGGUU_UUCUAGAUUCUGGCCCCU__________UUUUUCCACACUUUUCGGUGGUGUGG___GCCCACUCA_AAAGUUGUGGAUUAGAGCGUGUAAU .((((((((((..(......)..)))))))..................((((((((((..(((((.((.....)))))))...)))).))))))...)))........ (-17.65 = -19.65 + 2.00)



| Location | 18,169,988 – 18,170,082 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 97.46 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -31.12 |
| Energy contribution | -30.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18169988 94 - 27905053 GGCGCAUUCUUGCAGCCCUUGAUGCCCUCGGCAGUUGCCUUUCAUCUCUGCCUGGGAGAUCCUACUCGUCAGAGGGUU-UUCUGGAUUCUGGCCC ((((((....)))...((..((.(((((((((((.((.....))...)))))((((....)))).......)))))).-))..))......))). ( -32.30) >DroSec_CAF1 198216 94 - 1 GGCGCAUUCUUGCAGCCCUUGAUGCCCUCGGCAGUUGCCUUUCAUCUCUCCCUGGGAGAUCCUACUCGUCAGAGGGUU-UUCUAGAUUCUGGCCC ((((((....)))....((.((.(((((((((....)))....((((((.....))))))...........)))))).-.)).))......))). ( -31.10) >DroSim_CAF1 206062 94 - 1 GGCGCAUUCUUGCAGCCCUUGAUGCCCUCGGCAGUUGCCUUUCAUCUCUCCCUGGGAGAUCCUACUCGUCAGAGGGUU-UUCUAGAUUCUGGCCC ((((((....)))....((.((.(((((((((....)))....((((((.....))))))...........)))))).-.)).))......))). ( -31.10) >DroEre_CAF1 214076 94 - 1 GGCGCAUUCUUGCAGCCCUUGAUGCCCUCGGCAGUCGCCUUUCAUCUCUCCCUGGGAGAUCCUACUCGUCAGAGGGUU-UUCUGGAUUCUGGCCG ((((((....)))...((..((.(((((((((....)))....((((((.....))))))...........)))))).-))..))......))). ( -33.00) >DroYak_CAF1 208582 95 - 1 GGCGCAUUCUUGCAGCCCUUGAUGCCCUCGGCAGUCGCCUUUCAUCUCUCCCUGGGAGAUCCUACUCGUCAGAGGGUUUUUCUGGAUUCUGGCCC ((((((....)))...((..((.(((((((((....)))....((((((.....))))))...........))))))...)).))......))). ( -31.90) >consensus GGCGCAUUCUUGCAGCCCUUGAUGCCCUCGGCAGUUGCCUUUCAUCUCUCCCUGGGAGAUCCUACUCGUCAGAGGGUU_UUCUGGAUUCUGGCCC ((((((....)))....((.((.(((((((((....)))....((((((.....))))))...........))))))...)).))......))). (-31.12 = -30.88 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:55 2006