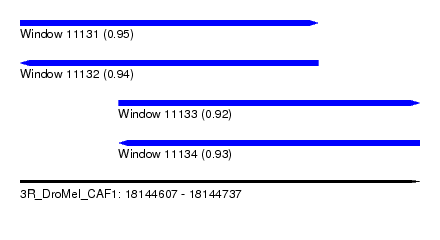
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,144,607 – 18,144,737 |
| Length | 130 |
| Max. P | 0.951322 |

| Location | 18,144,607 – 18,144,704 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -47.62 |
| Consensus MFE | -35.53 |
| Energy contribution | -36.82 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18144607 97 + 27905053 CGCCAGCGGCUCCGGUUGGUUGCAGCUGCUGGCGGCACUGCUGCAGCUGCUGCUAGAGGUUGCCUUCAUAGCAGCG---GCAACAGGAGCGAGUUGCACA----- (((((((((((.(((....))).)))))))))))((((((((.(.((((((((((((((....)))).))))))))---))....).))).)).)))...----- ( -51.90) >DroSec_CAF1 173230 97 + 1 CGCCAGCGGCUCCGGUUGGUUGCAGCUGCUGGCGGCACUGCUGCAGCUGCUGAUGGAGGUUGCCUUCAUAGCAGCG---GCAACAGGAGCAAGUUGCGCA----- (((((((((((.(((....))).)))))))))))((.(((.(((.(((((((.((((((...))))))))))))).---))).)))..((.....)))).----- ( -52.90) >DroEre_CAF1 184802 97 + 1 CGCCAGCGGCUCCGGCUGGUUGCAGCUGCUGGCGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCG---GCAACAGGAGCAAGUUGCGCA----- (((((((((((.(((....))).)))))))))))((.(((.(((.(((((((.((((((...))))))))))))).---))).)))..((.....)))).----- ( -54.00) >DroYak_CAF1 182408 97 + 1 UGCCAGCGGCUCCGGUUGGUUGCAGCUGCUGGCGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCG---GCAACAGGAGCAAGUUGCGCA----- .((((((.(((((.(((...(((((((((.....)))..))))))((((((((((((((....)))).))))))))---))))).)))))..)))).)).----- ( -51.60) >DroMoj_CAF1 224173 105 + 1 CGCCAGUGGCUCCAAGUGGUUGAUGCUGCUGGCGGCGCUGCUGCUGCUGCUGUUGACCGUUGCCUUCAUAGCAACGCCUGCAAAGAAACACAACAUCCACAUCCA ((((((..((..(((....)))..))..))))))..((.((....)).))(((((..((((((.......)))))).((....)).....))))).......... ( -34.60) >DroAna_CAF1 168117 97 + 1 GGCAGCCGCCUCCGGUUGAUUGCAGCUGCUGGCGGCACUGCUGCAACUGCUACUGGAUGUUGCUUUCAUAGCAGCG---GCAACUGGAAAGUGUUAAACG----- ((((((((((..((((((....))))))..))))))..))))((..((((((.((((.......))))))))))..---)).(((....)))........----- ( -40.70) >consensus CGCCAGCGGCUCCGGUUGGUUGCAGCUGCUGGCGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCG___GCAACAGGAGCAAGUUGCGCA_____ (((((((((((.(((....))).)))))))))))...(((.(((.(((((((.((((((...)))))))))))))....))).)))................... (-35.53 = -36.82 + 1.28)



| Location | 18,144,607 – 18,144,704 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -42.91 |
| Consensus MFE | -29.52 |
| Energy contribution | -30.80 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18144607 97 - 27905053 -----UGUGCAACUCGCUCCUGUUGC---CGCUGCUAUGAAGGCAACCUCUAGCAGCAGCUGCAGCAGUGCCGCCAGCAGCUGCAACCAACCGGAGCCGCUGGCG -----.........((((.((((.((---.(((((((.((.(....).))))))))).)).)))).)))).(((((((.(((.(........).))).))))))) ( -46.30) >DroSec_CAF1 173230 97 - 1 -----UGCGCAACUUGCUCCUGUUGC---CGCUGCUAUGAAGGCAACCUCCAUCAGCAGCUGCAGCAGUGCCGCCAGCAGCUGCAACCAACCGGAGCCGCUGGCG -----.((((.....))..(((((((---.(((((((((.(((...))).))).)))))).))))))).))(((((((.(((.(........).))).))))))) ( -47.10) >DroEre_CAF1 184802 97 - 1 -----UGCGCAACUUGCUCCUGUUGC---CGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCGCCAGCAGCUGCAACCAGCCGGAGCCGCUGGCG -----.((((.....))..(((((((---.((((((.((.(((...))).))..)))))).))))))).))(((((((.(((.(........).))).))))))) ( -46.40) >DroYak_CAF1 182408 97 - 1 -----UGCGCAACUUGCUCCUGUUGC---CGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCGCCAGCAGCUGCAACCAACCGGAGCCGCUGGCA -----.((((.....))..(((((((---.((((((.((.(((...))).))..)))))).))))))).)).((((((.(((.(........).))).)))))). ( -45.70) >DroMoj_CAF1 224173 105 - 1 UGGAUGUGGAUGUUGUGUUUCUUUGCAGGCGUUGCUAUGAAGGCAACGGUCAACAGCAGCAGCAGCAGCGCCGCCAGCAGCAUCAACCACUUGGAGCCACUGGCG ....((..((.(........)))..))(((((((((.(((.(....)..)))...((....)))))))))))(((((..((..(((....)))..))..))))). ( -37.50) >DroAna_CAF1 168117 97 - 1 -----CGUUUAACACUUUCCAGUUGC---CGCUGCUAUGAAAGCAACAUCCAGUAGCAGUUGCAGCAGUGCCGCCAGCAGCUGCAAUCAACCGGAGGCGGCUGCC -----.......((((.....(((((---.((((((((..............)))))))).)))))))))......((((((((..((....))..)))))))). ( -34.44) >consensus _____UGCGCAACUUGCUCCUGUUGC___CGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCGCCAGCAGCUGCAACCAACCGGAGCCGCUGGCG ...................(((((((....((((((..((.(....).))....)))))).)))))))....((((((.(((.(........).))).)))))). (-29.52 = -30.80 + 1.28)


| Location | 18,144,639 – 18,144,737 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -26.44 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18144639 98 + 27905053 CGGCACUGCUGCAGCUGCUGCUAGAGGUUGCCUUCAUAGCAGCGGCAACAGGAGCGAGUUGCACAGAGUACUCUGUU---CGAAAAAAAAAUUUGAUAAGA---- ...(.((.(((..((((((((((((((....)))).))))))))))..))).)).)..(((.(((((....))))).---)))..................---- ( -34.50) >DroSec_CAF1 173262 96 + 1 CGGCACUGCUGCAGCUGCUGAUGGAGGUUGCCUUCAUAGCAGCGGCAACAGGAGCAAGUUGCGCAGAGCACUCUGUC---GGAAA--AAAAUUAAAUAAGA---- (((((((.((((.(((((((.((((((...))))))))))))).(((((........))))))))))).....))))---)....--..............---- ( -35.40) >DroSim_CAF1 178181 96 + 1 CGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCGGCAACAGGAGCAAGUUGCACAGAGUACUCUGUC---GGAAA--AAAAUUAGGUAAGA---- ..((.(((.(((.(((((((.((((((...))))))))))))).))).)))..))...(((.(((((....))))))---))...--..............---- ( -34.20) >DroEre_CAF1 184834 95 + 1 CGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCGGCAACAGGAGCAAGUUGCGCAGUGUACUCUGCG---GGUAG---AAAUAAGUUGGGG---- ..(((((((....(((((((.((((((...))))))))))))).(((((........)))))))))))).(((.((.---..((.---...)).)).))).---- ( -40.70) >DroYak_CAF1 182440 98 + 1 CGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCGGCAACAGGAGCAAGUUGCGCAGUGUACUCUGUGUAGUGUAG---AAAGAAGUUAUGA---- ..((((((((.(.(((((((.((((((...)))))))))))))(....).).))))...(((((((......)))))))))))..---.............---- ( -38.20) >DroAna_CAF1 168149 98 + 1 CGGCACUGCUGCAACUGCUACUGGAUGUUGCUUUCAUAGCAGCGGCAACUGGAAAGUGUUAAACGGGAUACUCUGAG---G-AAG---AAAUCAGAAAAUAUAUU .((((((..(((..((((((.((((.......))))))))))..))).......))))))...........(((((.---.-...---...)))))......... ( -24.20) >consensus CGGCACUGCUGCAGCUGCUGCUGGAGGUUGCCUUCAUAGCAGCGGCAACAGGAGCAAGUUGCACAGAGUACUCUGUG___GGAAA___AAAUUAGAUAAGA____ ..((.(((.(((.(((((((.((((((...))))))))))))).))).)))..)).......((((......))))............................. (-26.44 = -27.05 + 0.61)

| Location | 18,144,639 – 18,144,737 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18144639 98 - 27905053 ----UCUUAUCAAAUUUUUUUUUCG---AACAGAGUACUCUGUGCAACUCGCUCCUGUUGCCGCUGCUAUGAAGGCAACCUCUAGCAGCAGCUGCAGCAGUGCCG ----....................(---.(((((....))))).)....((((.((((.((.(((((((.((.(....).))))))))).)).)))).))))... ( -32.60) >DroSec_CAF1 173262 96 - 1 ----UCUUAUUUAAUUUU--UUUCC---GACAGAGUGCUCUGCGCAACUUGCUCCUGUUGCCGCUGCUAUGAAGGCAACCUCCAUCAGCAGCUGCAGCAGUGCCG ----..............--....(---(.((((....))))))......((..(((((((.(((((((((.(((...))).))).)))))).))))))).)).. ( -32.20) >DroSim_CAF1 178181 96 - 1 ----UCUUACCUAAUUUU--UUUCC---GACAGAGUACUCUGUGCAACUUGCUCCUGUUGCCGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCG ----..............--.....---.(((((....))))).......((..(((((((.((((((.((.(((...))).))..)))))).))))))).)).. ( -31.60) >DroEre_CAF1 184834 95 - 1 ----CCCCAACUUAUUU---CUACC---CGCAGAGUACACUGCGCAACUUGCUCCUGUUGCCGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCG ----.............---.....---(((((......)))))......((..(((((((.((((((.((.(((...))).))..)))))).))))))).)).. ( -33.00) >DroYak_CAF1 182440 98 - 1 ----UCAUAACUUCUUU---CUACACUACACAGAGUACACUGCGCAACUUGCUCCUGUUGCCGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCG ----.............---.................(((((((((((........))))).((((((.((.(((...))).))..))))))....))))))... ( -29.40) >DroAna_CAF1 168149 98 - 1 AAUAUAUUUUCUGAUUU---CUU-C---CUCAGAGUAUCCCGUUUAACACUUUCCAGUUGCCGCUGCUAUGAAAGCAACAUCCAGUAGCAGUUGCAGCAGUGCCG ........((((((...---...-.---.))))))............((((.....(((((.((((((((..............)))))))).)))))))))... ( -23.74) >consensus ____UCUUAUCUAAUUU___CUUCC___CACAGAGUACUCUGCGCAACUUGCUCCUGUUGCCGCUGCUAUGAAGGCAACCUCCAGCAGCAGCUGCAGCAGUGCCG ..............................(((......)))........((..(((((((.((((((.((.(((...))).))..)))))).))))))).)).. (-22.85 = -23.10 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:44 2006