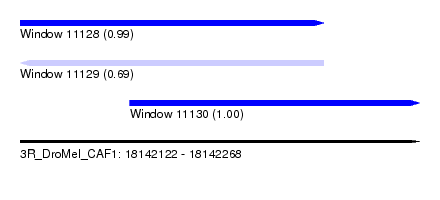
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,142,122 – 18,142,268 |
| Length | 146 |
| Max. P | 0.999867 |

| Location | 18,142,122 – 18,142,233 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -23.22 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18142122 111 + 27905053 CGCAUUCAUGUAAUGCACAUGCAAACAUUUGUGCUGAAAUGCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUAC---UACUACAACUACAGCUGCAGAUGUAG-- .((((..((((..(((....))).))))..)))).............((((((((((((.(((.((((((...........))).---.)))...)))))))))))))))....-- ( -31.60) >DroSec_CAF1 170769 114 + 1 CGCAUUCAUGUAAUGCACAUGCAAACAUUUGUGCUGAAAUGCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUACUACUACUACAACUACAGCUGCAGAUGUAG-- .((((..((((..(((....))).))))..)))).............((((((((((((..(((.....)))....((......))............))))))))))))....-- ( -31.40) >DroSim_CAF1 175690 114 + 1 CGCAUUCAUGUAAUGCACAUGCAAACAUUUGUGCUGAAAUGCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUACUACUACUACAACUACAGCUGCAGAUGUAG-- .((((..((((..(((....))).))))..)))).............((((((((((((..(((.....)))....((......))............))))))))))))....-- ( -31.40) >DroEre_CAF1 182392 105 + 1 CGCAUUCAUGUAAUGCACAUGCAAACAUUUGUGCUGAAAUGCAACUUGUCUGCAGCUGUCAGUCAGUUAGAUACACAGAUACUACUACUACU---------GCUGCAGAUGUAC-- .((((..((((..(((....))).))))..)))).............(((((((((.((.(((.(((...((......))...)))))))).---------)))))))))....-- ( -29.20) >DroAna_CAF1 165482 108 + 1 CGCAUUCAAGUAAUGCACAUGCAAACAUUUGUGCUGAAAUGCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAAAUACAC--------ACACAGACAAUCGCACAAGCGCCC .(((((.....)))))..............(((((....(((...(((((((.(((((.....))))).(.....)........--------...)))))))..)))..))))).. ( -22.50) >consensus CGCAUUCAUGUAAUGCACAUGCAAACAUUUGUGCUGAAAUGCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUACUACUACUACAACUACAGCUGCAGAUGUAG__ .(((((((......(((((..........))))))).))))).....((((((((((((..(((.....)))....((......))............))))))))))))...... (-23.22 = -24.90 + 1.68)

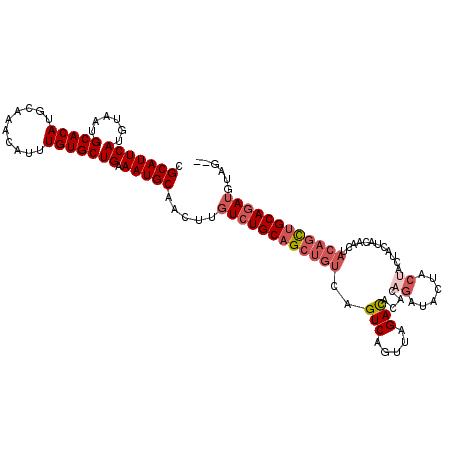

| Location | 18,142,122 – 18,142,233 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -23.77 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18142122 111 - 27905053 --CUACAUCUGCAGCUGUAGUUGUAGUA---GUAGUAUCUGUGUGUCUAACUGACUGACAGCUGCAGACAAGUUGCAUUUCAGCACAAAUGUUUGCAUGUGCAUUACAUGAAUGCG --((((..(((((((....)))))))..---))))...((((..(((.....)))..)))).((((((((...(((......)))....))))))))...(((((.....))))). ( -33.70) >DroSec_CAF1 170769 114 - 1 --CUACAUCUGCAGCUGUAGUUGUAGUAGUAGUAGUAUCUGUGUGUCUAACUGACUGACAGCUGCAGACAAGUUGCAUUUCAGCACAAAUGUUUGCAUGUGCAUUACAUGAAUGCG --((((..(((((((....)))))))..))))..((((..(((((((.....)))((...((((((((((...(((......)))....)))))))).)).))..))))..)))). ( -34.20) >DroSim_CAF1 175690 114 - 1 --CUACAUCUGCAGCUGUAGUUGUAGUAGUAGUAGUAUCUGUGUGUCUAACUGACUGACAGCUGCAGACAAGUUGCAUUUCAGCACAAAUGUUUGCAUGUGCAUUACAUGAAUGCG --((((..(((((((....)))))))..))))..((((..(((((((.....)))((...((((((((((...(((......)))....)))))))).)).))..))))..)))). ( -34.20) >DroEre_CAF1 182392 105 - 1 --GUACAUCUGCAGC---------AGUAGUAGUAGUAUCUGUGUAUCUAACUGACUGACAGCUGCAGACAAGUUGCAUUUCAGCACAAAUGUUUGCAUGUGCAUUACAUGAAUGCG --.....((((((((---------..((((((((((((....))).)).))).))))...))))))))......(((((.(((((((..((....)))))))......))))))). ( -27.90) >DroAna_CAF1 165482 108 - 1 GGGCGCUUGUGCGAUUGUCUGUGU--------GUGUAUUUGUGUGUCUAACUGACUGACAGCUGCAGACAAGUUGCAUUUCAGCACAAAUGUUUGCAUGUGCAUUACUUGAAUGCG (((((.((((((((.(((...(((--------.((((.((((..(((.....)))..)))).)))).)))....)))..)).)))))).)))))((((...((.....)).)))). ( -31.70) >consensus __CUACAUCUGCAGCUGUAGUUGUAGUAGUAGUAGUAUCUGUGUGUCUAACUGACUGACAGCUGCAGACAAGUUGCAUUUCAGCACAAAUGUUUGCAUGUGCAUUACAUGAAUGCG .......(((((((((((..........................(((.....)))..)))))))))))......(((((.(((((((..((....)))))))......))))))). (-23.77 = -24.65 + 0.88)



| Location | 18,142,162 – 18,142,268 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -13.52 |
| Energy contribution | -15.20 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18142162 106 + 27905053 GCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUAC---UACUACAACUACAGCUGCAGAUGUAG---UACAUAACCACCCGCACACCCACACACCCACACAC ...((((((((((((((((.(((.((((((...........))).---.)))...)))))))))))))))).))---).................................. ( -23.50) >DroSec_CAF1 170809 93 + 1 GCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUACUACUACUACAACUACAGCUGCAGAUGUAG---UACAUAACCAC----------------CCACACAC ...((((((((((((((((..(((.....)))....((......))............))))))))))))).))---)..........----------------........ ( -23.30) >DroSim_CAF1 175730 93 + 1 GCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUACUACUACUACAACUACAGCUGCAGAUGUAG---UACAUAACCAC----------------CCACACAC ...((((((((((((((((..(((.....)))....((......))............))))))))))))).))---)..........----------------........ ( -23.30) >DroEre_CAF1 182432 98 + 1 GCAACUUGUCUGCAGCUGUCAGUCAGUUAGAUACACAGAUACUACUACUACU---------GCUGCAGAUGUAC---UACAUACCC--CCAUACAUCCACACACCCACACAC .......(((((((((.((.(((.(((...((......))...)))))))).---------)))))))))....---.........--........................ ( -19.50) >DroAna_CAF1 165522 88 + 1 GCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAAAUACAC--------ACACAGACAAUCGCACAAGCGCCCUGGCAGACUCAC----------------AGAUACAC ......((((((.(((((((((((.....)))............--------...........(((....)))...)))))).))..)----------------)))))... ( -18.60) >consensus GCAACUUGUCUGCAGCUGUCAGUCAGUUAGACACACAGAUACUACUACUACUACAACUACAGCUGCAGAUGUAG___UACAUAACCAC________________CCACACAC .......((((((((((((..(((.....)))....((......))............)))))))))))).......................................... (-13.52 = -15.20 + 1.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:41 2006