
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,139,767 – 18,139,863 |
| Length | 96 |
| Max. P | 0.630532 |

| Location | 18,139,767 – 18,139,863 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -19.18 |
| Energy contribution | -20.44 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
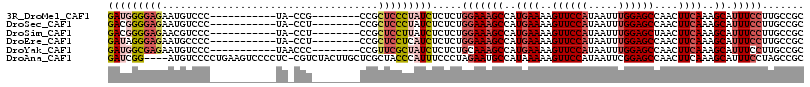
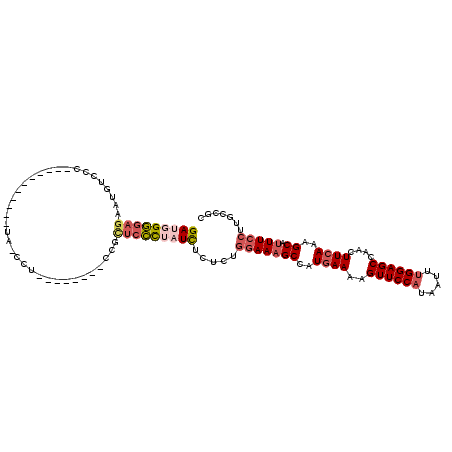
>3R_DroMel_CAF1 18139767 96 - 27905053 GAUGGGGAGAAUGUCCC-----------UA-CCG--------CCGCUCCCUAUCUCUCUGGAAAGCCAUGAAAAGUUCCAUAAUUUGGAGCCAACUUCAAAGCAUUUCCUUGCCGC (((((((((...((...-----------..-..)--------)..))))))))).....(((((((..((((..((((((.....))))))....))))..)).)))))....... ( -31.00) >DroSec_CAF1 168384 96 - 1 GACGGGGAGAAUGUCCC-----------UA-CCU--------CCGCUCCCUAUCUCUCUGGAAAGCCAUGAAAAGUUCCAUAAUUUGGAGCCAACUUCAAAGCAUUUCCUUGCCGC ((..(((((...((...-----------.)-)..--------...)))))..)).....(((((((..((((..((((((.....))))))....))))..)).)))))....... ( -26.60) >DroSim_CAF1 173319 96 - 1 GACGGGGAGAACGUCCC-----------UA-CCU--------CCGCUCCUUAUCUCUCUGGAAAGCCAUGAAAAGUUCCAUAAUUUGGAGCUAACUUCAAAGCAUUUCCUUGCCGC ((..(((((...((...-----------.)-)..--------...)))))..)).....(((((((..((((.(((((((.....)))))))...))))..)).)))))....... ( -26.50) >DroEre_CAF1 180064 96 - 1 GAUAGGGAGAAUGCCCC-----------UA-CCU--------CCGCUCCUCAUCUCUCUGGAAAGCCAUGAAAAGUUCCAUAAUUUGGAGCCAACUUCAAAGCAUUUCCUUGCCGC (((..((((........-----------..-...--------...))))..))).....(((((((..((((..((((((.....))))))....))))..)).)))))....... ( -24.09) >DroYak_CAF1 177407 97 - 1 GAUGGCGAGAAUGUCCC-----------UAACCC--------CCGUUCGCUAUCUCUCUGCAAAGCCAUGAAAAGUUCCAUAAUUUGGAGCCAACUUCAAAGCAUUUCCUUGCCGC ...((((((........-----------......--------......((.........))...((..((((..((((((.....))))))....))))..)).....)))))).. ( -22.50) >DroAna_CAF1 163545 111 - 1 GAUCGG----AUGUCCCCUGAAGUCCCCUC-CGUCUACUUGCUCGCUACCCAUUUCCCUAGAAUGCCAUAAAAAGUUCCAUAAUUCGGAGCCAACUUCAAAGCAUUUCCUAGCCGC ....((----(.((.(..((((((...(((-((.......(((.......(((((.....)))))........))).........)))))...))))))..).)).)))....... ( -18.35) >consensus GAUGGGGAGAAUGUCCC___________UA_CCU________CCGCUCCCUAUCUCUCUGGAAAGCCAUGAAAAGUUCCAUAAUUUGGAGCCAACUUCAAAGCAUUUCCUUGCCGC (((((((((....................................))))))))).....(((((((..((((..((((((.....))))))....))))..)).)))))....... (-19.18 = -20.44 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:37 2006