
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,137,579 – 18,137,717 |
| Length | 138 |
| Max. P | 0.991920 |

| Location | 18,137,579 – 18,137,679 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18137579 100 + 27905053 ACAAUUUCUAUAAGUGAGUGGUAUGUGGUUUCUGUUAUCCUGUAUAUUCGGUCUAAGCAGCAGCGAUGCUGCUUAGGGCCGAAUCCUAGGGAUGCUUCGU .((....((((......))))....))........((((((....(((((((((((((((((....))))))))).))))))))....))))))...... ( -35.10) >DroSec_CAF1 166176 97 + 1 ACAAUUUCUAUAAGUGAGUGAUGUGUGGUUUUUGUUAUCCUGUAUAUUCGGUCUAAGCAGCAGCG---CUGCUUAGGGCCGAAUCCUAGGGAUGCUUUGU ((((...(((((.((.....)).)))))...))))((((((....((((((((((((((((...)---))))))).))))))))....))))))...... ( -33.30) >DroSim_CAF1 171079 97 + 1 ACAAUUUCUAUAAGUGAGUGGUGUGUGGUUUUUGUUAUCCUGUAUAUUCGGUCUAAGCAGCAGCG---CUGCUUAGGGCCGAAUCCUAGGGAUGCUUUGU ((((...(((((..(.....)..)))))...))))((((((....((((((((((((((((...)---))))))).))))))))....))))))...... ( -32.30) >DroEre_CAF1 177819 94 + 1 ACAAUUUCUAUAAGUGAGU-AUGU--GUCUUCUGUAUUCCUGGGUAUUGGUACUAAGCAGCAGCG---CUGCUUAGGGCCGAAUCCUAGGGAUGCUUCGU (((..(((.......))).-.)))--.......(((((((((((..((((..(((((((((...)---))))))))..))))..)))))))))))..... ( -38.80) >DroYak_CAF1 175113 96 + 1 ACAAUUUCUAUAAGUGAGU-AUGUGUGUUUUCUGUUAUCCUGCAUAUUGGUACUAAGCAGCAGCG---CUGCUUAGGGCCGAAUCCUAGGGAUGCUUCGU (((..(((.......))).-...))).........((((((.....((((..(((((((((...)---))))))))..))))......))))))...... ( -27.30) >DroPer_CAF1 194206 89 + 1 --AAUGCCUGUG----UGUUGAUUGGGGGUUCGAUUGUUUAG--CGUCGAUCUUAAGCAGCAGCG---CUGCUUAGGGCCGUGUCCUAGGGAUGCUUCGU --....((((..----.......))))((((((((((.....--...))))).((((((((...)---))))))))))))((((((...))))))..... ( -26.90) >consensus ACAAUUUCUAUAAGUGAGUGAUGUGUGGUUUCUGUUAUCCUGUAUAUUCGGUCUAAGCAGCAGCG___CUGCUUAGGGCCGAAUCCUAGGGAUGCUUCGU ...................................((((((........(.(((((((((........))))))))).).........))))))...... (-14.90 = -15.30 + 0.39)


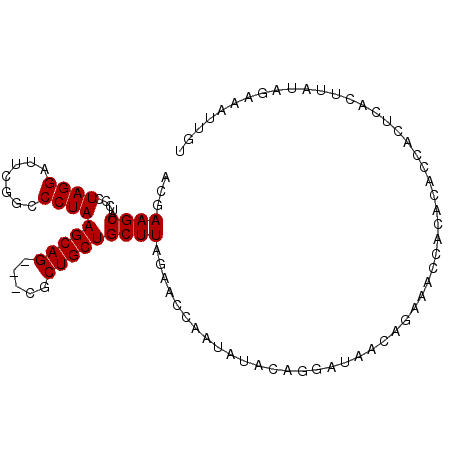
| Location | 18,137,579 – 18,137,679 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18137579 100 - 27905053 ACGAAGCAUCCCUAGGAUUCGGCCCUAAGCAGCAUCGCUGCUGCUUAGACCGAAUAUACAGGAUAACAGAAACCACAUACCACUCACUUAUAGAAAUUGU .......((((.....((((((..((((((((((....)))))))))).)))))).....)))).................................... ( -27.80) >DroSec_CAF1 166176 97 - 1 ACAAAGCAUCCCUAGGAUUCGGCCCUAAGCAG---CGCUGCUGCUUAGACCGAAUAUACAGGAUAACAAAAACCACACAUCACUCACUUAUAGAAAUUGU .......((((.....((((((..((((((((---(...))))))))).)))))).....)))).................................... ( -25.30) >DroSim_CAF1 171079 97 - 1 ACAAAGCAUCCCUAGGAUUCGGCCCUAAGCAG---CGCUGCUGCUUAGACCGAAUAUACAGGAUAACAAAAACCACACACCACUCACUUAUAGAAAUUGU .......((((.....((((((..((((((((---(...))))))))).)))))).....)))).................................... ( -25.30) >DroEre_CAF1 177819 94 - 1 ACGAAGCAUCCCUAGGAUUCGGCCCUAAGCAG---CGCUGCUGCUUAGUACCAAUACCCAGGAAUACAGAAGAC--ACAU-ACUCACUUAUAGAAAUUGU ..........(((.(((((.((..((((((((---(...)))))))))..))))).)).)))............--....-................... ( -22.90) >DroYak_CAF1 175113 96 - 1 ACGAAGCAUCCCUAGGAUUCGGCCCUAAGCAG---CGCUGCUGCUUAGUACCAAUAUGCAGGAUAACAGAAAACACACAU-ACUCACUUAUAGAAAUUGU .....((((((...)))...((..((((((((---(...)))))))))..))....))).....................-................... ( -20.60) >DroPer_CAF1 194206 89 - 1 ACGAAGCAUCCCUAGGACACGGCCCUAAGCAG---CGCUGCUGCUUAAGAUCGACG--CUAAACAAUCGAACCCCCAAUCAACA----CACAGGCAUU-- .((((((.(((...)))..((..(.(((((((---(...)))))))).)..))..)--))......)))...............----..........-- ( -18.30) >consensus ACGAAGCAUCCCUAGGAUUCGGCCCUAAGCAG___CGCUGCUGCUUAGAACCAAUAUACAGGAUAACAGAAACCACACACCACUCACUUAUAGAAAUUGU ...((((.....((((.......))))(((((.....)))))))))...................................................... (-11.77 = -11.77 + 0.00)


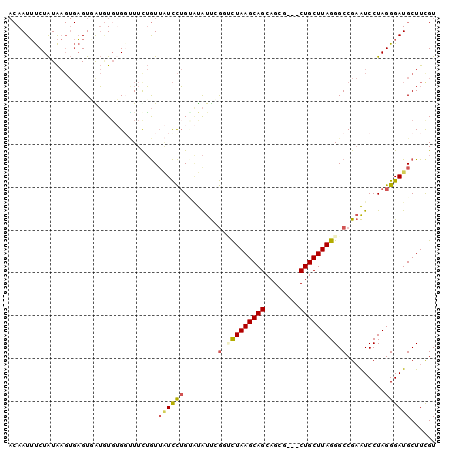
| Location | 18,137,610 – 18,137,717 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.27 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -26.36 |
| Energy contribution | -26.45 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
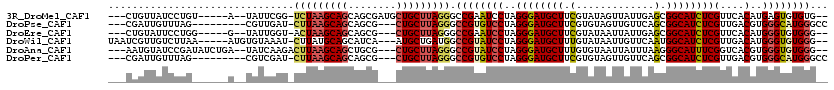
>3R_DroMel_CAF1 18137610 107 + 27905053 ---CUGUUAUCCUGU-----A--UAUUCGG-UCUAAGCAGCAGCGAUGCUGCUUAGGGCCGAAUCCUAGGGAUGCUUCGUAUAGUUAUUGAGCGGCAUCUCGUUCACAUGAGUGUGUG-- ---....((((((..-----.--.((((((-(((((((((((....))))))))).))))))))....)))))).....((((.(((((((((((....))))))).)))).))))..-- ( -43.50) >DroPse_CAF1 168390 104 + 1 ---CGAUUGUUUAG---------CGUUGAU-CUUAAGCAGCAGCG---CUGCUUAGGGCCGUGUCCUAGGGAUGCUUCGUGUAGUUGUUCAGCGGCAUCUCGUUGACGUGGGCAUGGGCC ---.(((((.....---------...))))-).((((((((...)---)))))))((.((((((((..((((((((.(.((........))).))))))))(....)..)))))))).)) ( -39.40) >DroEre_CAF1 177847 104 + 1 ---CUGUAUUCCUGG-----G--UAUUGGU-ACUAAGCAGCAGCG---CUGCUUAGGGCCGAAUCCUAGGGAUGCUUCGUAUAAUUAUUGAGCGGCAUCUCGUUCACAUGGGUGUGGG-- ---..((((((((((-----(--..((((.-.(((((((((...)---))))))))..))))..)))))))))))..(.((((.(((((((((((....))))))).)))).)))).)-- ( -46.50) >DroWil_CAF1 183791 109 + 1 UAAUCGUUGUCUUAA-----AUGUGUAAAU-CUUAUGCAGCAUCA---AUGCUGAUGGCCGUAUCCUAGGGAUGCUUUGUAUAAUUGUUCAAUGGCAUCUCGUUGACAUGGGUGUGGG-- ....((((......)-----))).......-(((((((..(((((---....))))).((((.((...(((((((((((.((....)).))).))))))))...)).)))))))))))-- ( -26.00) >DroAna_CAF1 161496 110 + 1 ---AAUGUAUCCGAUAUCUGA--UAUCAAGACUUAAGCAGCUGCG---CUGCUUAGGGCCGUAUCCUAGGGAUGCUUUGUGUAAUUAUUUAAGGGCAUUUCGGUCACGUGGGUGUGGG-- ---.......((.((((((..--........((((((((((...)---)))))))))..(((..((..(..(((((((.............)))))))..)))..))).)))))).))-- ( -34.52) >DroPer_CAF1 194231 104 + 1 ---CGAUUGUUUAG---------CGUCGAU-CUUAAGCAGCAGCG---CUGCUUAGGGCCGUGUCCUAGGGAUGCUUCGUGUAGUUGUUCAGCGGCAUCUCGUUGACGUGGGCAUGGGCC ---.(((((.....---------...))))-).((((((((...)---)))))))((.((((((((..((((((((.(.((........))).))))))))(....)..)))))))).)) ( -41.50) >consensus ___CGGUUGUCCUG______A__UAUUGAU_CUUAAGCAGCAGCG___CUGCUUAGGGCCGUAUCCUAGGGAUGCUUCGUAUAAUUAUUCAGCGGCAUCUCGUUCACAUGGGUGUGGG__ ...............................(((((((((........))))))))).((((((((..((((((((.(.............).))))))))(....)..))))))))... (-26.36 = -26.45 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:35 2006