
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,135,949 – 18,136,077 |
| Length | 128 |
| Max. P | 0.999951 |

| Location | 18,135,949 – 18,136,046 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.46 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -33.58 |
| Energy contribution | -33.67 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.80 |
| SVM RNA-class probability | 0.999951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18135949 97 - 27905053 CCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAAUUGAAACAUUUAGCGGCUACGUGAGUGCGUU .......((((((((((..(((((((....)))))))))))))))))((....((((((.(((...........))).))))))((....))))... ( -36.50) >DroSec_CAF1 164565 97 - 1 CCAUUUCGGUGCUGCAACUGCGUUUAUUAUUAAAUGUCUGUGGCGCCGCGUUGAGCUGCCAGAAAAUUGAAACAUUUAGCGGCUACGUGAGUGCAUU .......(((((..((...(((((((....))))))).))..)))))((....((((((.(((...........))).))))))((....))))... ( -31.10) >DroSim_CAF1 169447 97 - 1 CCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGUGGCGCCGCGUUGAGCUGCCAGAAAAUUGAAACAUUUAGCGGCUACGUGAGUGCGUU .......(((((..(((..(((((((....))))))))))..)))))((....((((((.(((...........))).))))))((....))))... ( -34.00) >DroEre_CAF1 176149 97 - 1 CCAUUUCGGCGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAAUUGAAACAUUUAGCGGCUACGUGAGUGCGUU .......((((((((((..(((((((....)))))))))))))))))((....((((((.(((...........))).))))))((....))))... ( -38.40) >DroYak_CAF1 173417 97 - 1 CCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAAUUGAAACAUUUAGCGGCUACGUGAGUGCGUU .......((((((((((..(((((((....)))))))))))))))))((....((((((.(((...........))).))))))((....))))... ( -36.50) >DroAna_CAF1 159845 93 - 1 ----UUUGGCGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCACGUUGAGCUGCCAGAAAAUUGAAACAUUUACCCGCUACGUGAGUGCGUU ----..(((((((((((..(((((((....))))))))))))))))))................................(((.((....))))).. ( -33.30) >consensus CCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAAUUGAAACAUUUAGCGGCUACGUGAGUGCGUU .......((((((((((..(((((((....)))))))))))))))))((((..((((((.(((...........))).))))))((....)))))). (-33.58 = -33.67 + 0.09)

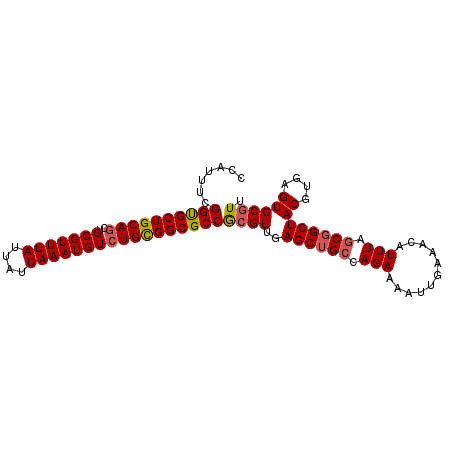

| Location | 18,135,980 – 18,136,077 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -29.99 |
| Energy contribution | -31.22 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18135980 97 - 27905053 AAAGCCAGCGAGACAUCCGGGAAAGUUCUGGCCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAA ..(((((((((((...((((((...))))))...))))((((((((((..(((((((....)))))))))))))))))..)))).)))......... ( -39.40) >DroSec_CAF1 164596 97 - 1 AAAGCCAGCGAGACAUCCGGGAAAGUUCUGGCCAUUUCGGUGCUGCAACUGCGUUUAUUAUUAAAUGUCUGUGGCGCCGCGUUGAGCUGCCAGAAAA ..(((((((((((...((((((...))))))...))))(((((..((...(((((((....))))))).))..)))))..)))).)))......... ( -34.00) >DroSim_CAF1 169478 97 - 1 AAAGCCAGCGAGACAUCCGGGAAAGUUCUGGCCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGUGGCGCCGCGUUGAGCUGCCAGAAAA ..(((((((((((...((((((...))))))...))))(((((..(((..(((((((....))))))))))..)))))..)))).)))......... ( -36.90) >DroEre_CAF1 176180 96 - 1 AGAGCCAGGGAGACAUCCGGGAAAGUUCUG-CCAUUUCGGCGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAA ....((..(((....))).))....(((((-.((....((((((((((..(((((((....)))))))))))))))))((.....)))).))))).. ( -38.00) >DroYak_CAF1 173448 97 - 1 AAAGCCAGGGAGACAGCCGGGAAAGUUCUGGCCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAA ....((.((.......)).))....(((((((......((((((((((..(((((((....)))))))))))))))))((.....)).))))))).. ( -39.70) >DroAna_CAF1 159876 88 - 1 AAAGCCAUACAGACAUCCGGAAAAAC---------UUUGGCGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCACGUUGAGCUGCCAGAAAA .................(((...(((---------..(((((((((((..(((((((....)))))))))))))))))).)))...)))........ ( -30.10) >consensus AAAGCCAGCGAGACAUCCGGGAAAGUUCUGGCCAUUUCGGUGCUGCAGCUGCGUUUAUUAUUAAAUGUCUGCGGCGCCGCGUUGAGCUGCCAGAAAA .........................(((((((......((((((((((..(((((((....)))))))))))))))))((.....)).))))))).. (-29.99 = -31.22 + 1.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:31 2006