
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,131,650 – 18,131,782 |
| Length | 132 |
| Max. P | 0.980414 |

| Location | 18,131,650 – 18,131,753 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.48 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.45 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
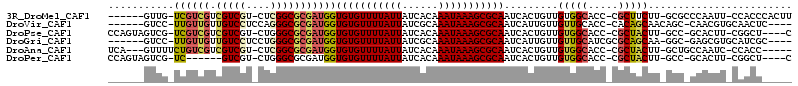
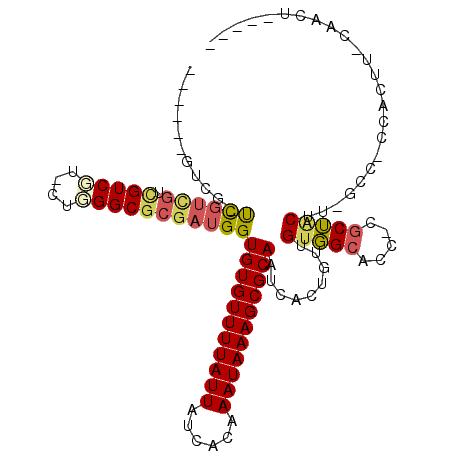
>3R_DroMel_CAF1 18131650 103 - 27905053 ------GUUG-UCGUCGUCGUCGU-CUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUUCUU-GCGCCCAAUU-CCACCCACUU ------...(-(((.(((((....-..)))))))))(((((((((((((......)))))))))).......((((.(((...-.((.....-))))))))).-..)))..... ( -28.80) >DroVir_CAF1 176880 101 - 1 ------GUCC-UUGUUGUUGUCCUCCAGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCACC-CACAGCAACAGC-CAACGUGCAACUC---- ------....-..(((((.((((....))))(((.((((((((((((((......))))))))))......((((((((....-..))))))))))-)).))))))))..---- ( -34.90) >DroPse_CAF1 162523 104 - 1 CCAGUAGUCG-UCGUCGUCGUCGU-CUGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUACUU-GCC-GCACUU-CGGCU----C .((((.....-((((((.((((..-...))))))))))(((((((((((......)))))))))))...))))..(((((...-.)))))..-(((-(.....-)))).----. ( -35.40) >DroGri_CAF1 177594 101 - 1 ------GUCC-UUGUUGUUGUCCUCCUGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCAUCGCGCAGCAA-GGC-GAGCGUGCAUCGC---- ------(.((-(((((((.((((....))))((((((.(((((((((((......)))))))))))..((....))...)))))))))))))-)))-..(((.....)))---- ( -38.40) >DroAna_CAF1 155410 102 - 1 UCA---GUUUUCUGUCGUCGUCGU-CUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUACUU-GCUGCCAAUC-CCACC----- .((---((...((((((.(((((.-..)))))))))))(((((((((((......)))))))))))...))))...(((((..-.((.....-)))))))...-.....----- ( -31.10) >DroPer_CAF1 188354 98 - 1 CCAGUAGUCG-UC------GUCGU-CUGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUACUU-GCC-GCACUU-CGGCU----C .((((..(((-((------(.(((-....)))))))))(((((((((((......)))))))))))...))))..(((((...-.)))))..-(((-(.....-)))).----. ( -34.00) >consensus ______GUCG_UCGUCGUCGUCGU_CUGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC_CGCUACUU_GCC_CCACUU_CAACU_____ ...........((((((.(((((....)))))))))))(((((((((((......))))))))))).........(((((.....)))))........................ (-22.50 = -22.45 + -0.05)

| Location | 18,131,681 – 18,131,782 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
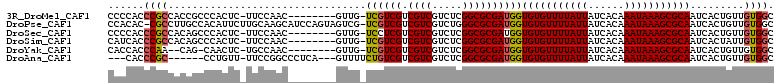
>3R_DroMel_CAF1 18131681 101 - 27905053 CCCCACCCGCCACCGCCCACUC-UUCCAAC--------GUUG-UCGUCGUCGUCGUCUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGC ........(((((.((......-.......--------....-((((((.(((((...)))))))))))(((((((((((......)))))))))))......)).))))) ( -30.80) >DroPse_CAF1 162549 109 - 1 CCACAC-CGCCUUGCCACAUUCUUGCAAGCAUCCAGUAGUCG-UCGUCGUCGUCGUCUGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGC (((((.-.((.((((.........))))))...((((.....-((((((.((((.....))))))))))(((((((((((......)))))))))))...)))).))))). ( -34.60) >DroSec_CAF1 160318 101 - 1 CCCCACCCGCCACAGCCCACUC-UUCCAAC--------GUUG-UCCUCGUCGUCGUCUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGC ........((((((((......-.......--------....-...((((((.(((....)))))))))(((((((((((......)))))))))))......)))))))) ( -32.40) >DroSim_CAF1 165188 101 - 1 CAUCACCCGCCACAGCCCACUC-UUCCAAC--------GUUG-UCGUCGUCGUCGUCUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUAUUGUGGC ........(((((((.......-.......--------....-((((((.(((((...)))))))))))(((((((((((......))))))))))).......))))))) ( -31.60) >DroYak_CAF1 168913 98 - 1 CACCACCCAA--CAG-CAACUC-UGCCAAC--------GUUG-UCGUCGUCGUCGUCUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGC .....(((((--(((-......-(((..((--------(..(-(((.(((((......)))))))))..)))((((((((......)))))))))))....)))))).)). ( -31.90) >DroAna_CAF1 155436 98 - 1 ---CACCCGC------CCUGUU-UUCCGGCCCUCA---GUUUUCUGUCGUCGUCGUCUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGC ---.......------......-.....((((.((---((...((((((.(((((...)))))))))))(((((((((((......)))))))))))...))))..).))) ( -27.90) >consensus CCCCACCCGCCACAGCCCACUC_UUCCAAC________GUUG_UCGUCGUCGUCGUCUCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGC ......((((.................................((((((.((((.....))))))))))(((((((((((......))))))))))).........)))). (-23.40 = -23.48 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:29 2006