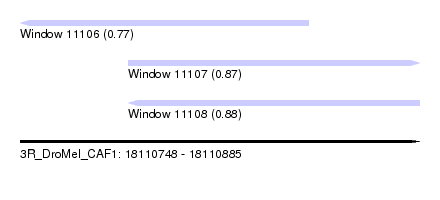
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,110,748 – 18,110,885 |
| Length | 137 |
| Max. P | 0.882866 |

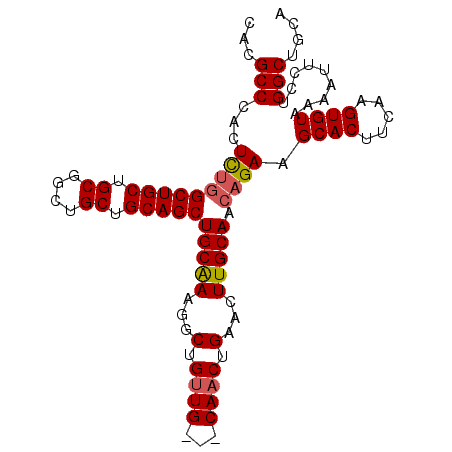
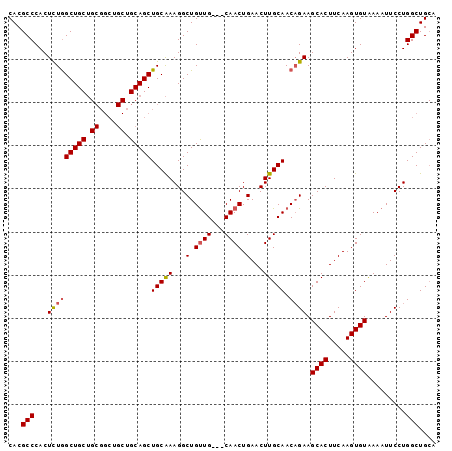
| Location | 18,110,748 – 18,110,847 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18110748 99 - 27905053 CACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGCUGCAGCAACUGAACUUGCAACAGAAGCACUUCAAGUGUAAAAUUCCUGGCUGCA ...(((...((((..(((..(((.(((((((((.((....)).))))))))).))).....))).)))).((((.....)))).........))).... ( -38.90) >DroSec_CAF1 141592 96 - 1 CACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGUUG---CAACUGAACUUGCAACAGAAGCACUUCAAGUGUAAAAUUCCUGGCUGCA ...(((.......((.(((((((...))))))).))..(((((((((---(((......)))))))))..((((.....))))......)))))).... ( -35.00) >DroSim_CAF1 146318 96 - 1 CACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGUUG---CAACUGAACUUGCAACUGAAGCACUUCAAGUGUAAAAUUCCUGGCUGCA ...(((......((((..((((((((...))))))))..))))((((---(((......)))))))....((((.....)))).........))).... ( -32.30) >DroEre_CAF1 151519 85 - 1 CACGCCCACUUUAGCUGC---------UGCAGCUGCGAAGGCUGUUG---CAACUGAACUUGCAACAGAAGCACUUCAAGUGUGAAAUUCCUGGCUG-- ...((((((((((((((.---------..)))))).((((.((((((---(((......))))))))).....)))))))))..........)))..-- ( -27.30) >consensus CACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGUUG___CAACUGAACUUGCAACAGAAGCACUUCAAGUGUAAAAUUCCUGGCUGCA ...(((...(((((((((.((....)).)))))(((((...(.((((...)))).)...))))).)))).((((.....)))).........))).... (-23.70 = -24.20 + 0.50)

| Location | 18,110,785 – 18,110,885 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -35.54 |
| Energy contribution | -36.23 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
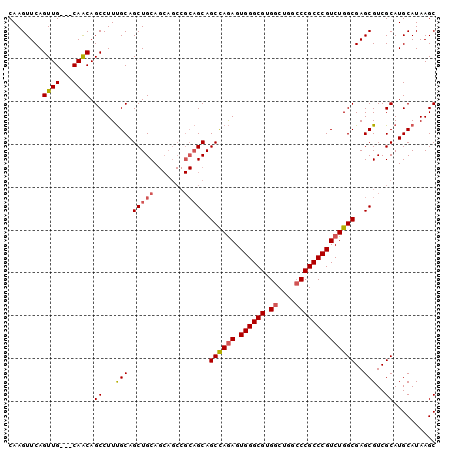
>3R_DroMel_CAF1 18110785 100 + 27905053 CAAGUUCAGUUGCUGCAGCAGCCUUUGCAGCUGCAGCAGCCGCAGCAGCCAGAGUGGGCGUGGCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGC ........(((((((((((.((....)).))))))))))).(((((.((((((.((((((.((....))))))))))))))(.....))).)))...... ( -53.60) >DroSec_CAF1 141629 97 + 1 CAAGUUCAGUUG---CAACAGCCUUUGCAGCUGCAGCAGCCGCAGCAGCCAGAGUGGGCGUGGCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGC ......((((((---(((......)))))))))..(((((.((.((.((((((.((((((.((....))))))))))))))..)))).)).)))...... ( -46.90) >DroSim_CAF1 146355 97 + 1 CAAGUUCAGUUG---CAACAGCCUUUGCAGCUGCAGCAGCCGCAGCAGCCAGAGUGGGCGUGGCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGC ......((((((---(((......)))))))))..(((((.((.((.((((((.((((((.((....))))))))))))))..)))).)).)))...... ( -46.90) >DroEre_CAF1 151554 88 + 1 CAAGUUCAGUUG---CAACAGCCUUCGCAGCUGCA---------GCAGCUAAAGUGGGCGUGGCUGGGCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGC ...(((((((((---(..........))))))).)---------)).(((...((((((((.((..(((.....)))..))..))))).))).....))) ( -30.70) >consensus CAAGUUCAGUUG___CAACAGCCUUUGCAGCUGCAGCAGCCGCAGCAGCCAGAGUGGGCGUGGCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGC ........((((...)))).((...(((.(((((.......))))).((((((.((((((.((....))))))))))))))..)))..)).......... (-35.54 = -36.23 + 0.69)

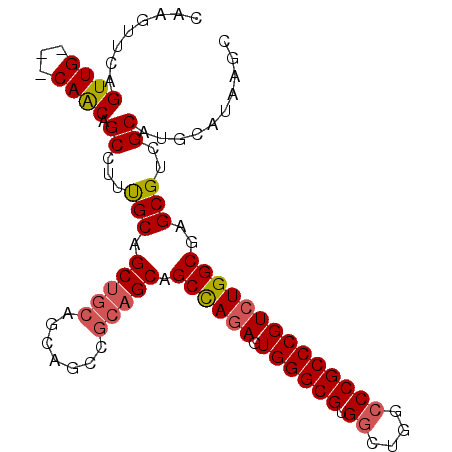
| Location | 18,110,785 – 18,110,885 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -35.25 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
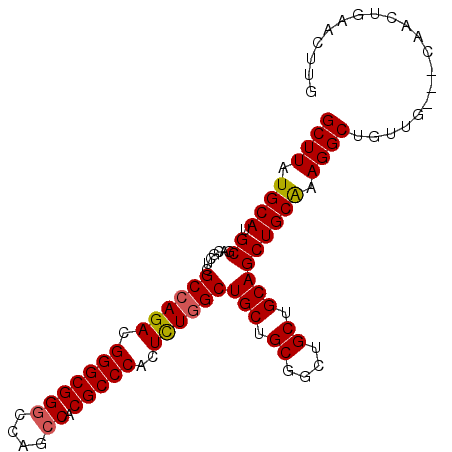
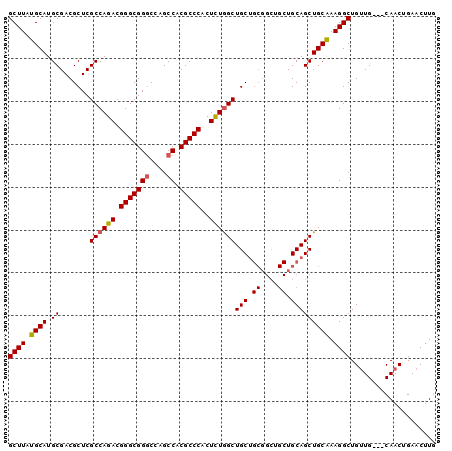
>3R_DroMel_CAF1 18110785 100 - 27905053 GCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGCCACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGCUGCAGCAACUGAACUUG ((....)).......((..((((((.(((((((....)).)))))..)))))).))..(((.(((((((((.((....)).))))))))).)))...... ( -50.20) >DroSec_CAF1 141629 97 - 1 GCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGCCACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGUUG---CAACUGAACUUG ((....)).(((((((((.((((((.(((((((....)).)))))..))))))...((((((((...))))))))..))).))))---)).......... ( -47.00) >DroSim_CAF1 146355 97 - 1 GCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGCCACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGUUG---CAACUGAACUUG ((....)).(((((((((.((((((.(((((((....)).)))))..))))))...((((((((...))))))))..))).))))---)).......... ( -47.00) >DroEre_CAF1 151554 88 - 1 GCUUAUGCAUGCGACGCUCGCCAGACGGGCGGCCCAGCCACGCCCACUUUAGCUGC---------UGCAGCUGCGAAGGCUGUUG---CAACUGAACUUG ((....))..(((..(((((.....)))))(((...))).)))....(((((.(((---------.((((((.....)))))).)---)).))))).... ( -31.10) >consensus GCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGCCACGCCCACUCUGGCUGCUGCGGCUGCUGCAGCUGCAAAGGCUGUUG___CAACUGAACUUG ((((.((((.((.......((((((.(((((((....)).)))))..))))))(((.((....)).))))))))).)))).................... (-35.25 = -35.50 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:20 2006