| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,110,519 – 18,110,635 |
| Length | 116 |
| Max. P | 0.530044 |

| Location | 18,110,519 – 18,110,635 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -14.00 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
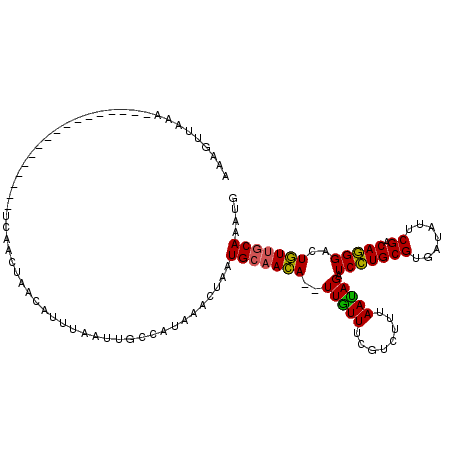
>3R_DroMel_CAF1 18110519 116 + 27905053 AAAAUUAGAGGAAAAGUUUUAGCAAAUCACCUACCAUUUAUUUGCCAUAAACUAAUGCAACA--UUGUUUUGUCUUUAAUAGAUCCUGCGUGAUAUUCGACAGGGACUGUUGCAAAUG ..............(((((..((((((............))))))...)))))..(((((((--..(((..((((.....))))((((((.......)).)))))))))))))).... ( -22.40) >DroSec_CAF1 141378 104 + 1 AAAGUUAAAGGA------------AAUCACCUACCAUUUAUUUGCCAUAAACUAAUGCAACA--UUGUUUCGUCUUUAAUAGAUCCUGCGUGAUAUUCGACAGGGACUGUUGCAAAUG ........(((.------------.....)))......(((((((...((((.((((...))--)))))).......(((((.(((((((.......)).))))).)))))))))))) ( -18.70) >DroSim_CAF1 146104 104 + 1 AAAGUUAAAGGA------------AAUCACCUACCAUUUAUUUGCCAUAAACUAAUGCAACA--UUGUUUCGUCUUUAACAGAUCCUGCGUGAUAUUCGACAGGGACUGUUGCAAAUG ........(((.------------.....)))......(((((((...((((.((((...))--)))))).......(((((.(((((((.......)).))))).)))))))))))) ( -20.60) >DroEre_CAF1 151323 91 + 1 AA------A-----------------UGAAGUAAA--CUAAUUCCCCUAAACUAAUGCAACA--UUGUUUCAUCUUUAAUAGAUCCUGCGUGAUAUUCGACAGGGACUGUUGCAAAUG ..------.-----------------.........--..................(((((((--........(((.....)))(((((((.......)).)))))..))))))).... ( -16.30) >DroYak_CAF1 149280 99 + 1 AACCCCAUA-----------------UUGAGUAACAUUUAAUUCACAUAAACUAAUGCAACA--UUGUUUCAUCUUUAAUAGAUCCUGCGUGAUAUUCGACAGGGACUGUUGCAAAUG .........-----------------.(((((........)))))..........(((((((--........(((.....)))(((((((.......)).)))))..))))))).... ( -17.90) >DroAna_CAF1 137057 93 + 1 AAAGCAUUA-----------------CUGAGUUACGU---AU-----UCGACUAAUGGCAUAAUUUAUUUUGUCCUCAAUAGAUCUUGCGUGACAUCCGACAAGGACUAUUACAAAUG ....((((.-----------------....(((((((---(.-----..(((((..((.((((......))))))....))).)).)))))))).(((.....)))........)))) ( -16.70) >consensus AAAGUUAAA_________________UCAACUAACAUUUAAUUGCCAUAAACUAAUGCAACA__UUGUUUCGUCUUUAAUAGAUCCUGCGUGAUAUUCGACAGGGACUGUUGCAAAUG .......................................................(((((((..(((((........))))).(((((((.......)).)))))..))))))).... (-14.00 = -13.50 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:17 2006