
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,107,039 – 18,107,131 |
| Length | 92 |
| Max. P | 0.689447 |

| Location | 18,107,039 – 18,107,131 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18107039 92 + 27905053 UUUUGACUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGCGGCUGUUGCUUUUGAGCACGUUUACCUGUAAUGGCAUGCCACCAGCAACAA--------- .......(((....))).((((.((((.....))))..))))..(((((((..((.(((.(((..........))).)))))..))))))).--------- ( -21.60) >DroVir_CAF1 151995 83 + 1 CUGCCGUUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGCAACCGUUGCUGCCGAGCACGUUUACCUGCAAUGGCA-----GCAGCA------------- .....(((((....)))))(((.((((.....))))..)))....((((((((((.(((.(....).)))..)))))-----))))).------------- ( -30.10) >DroPse_CAF1 141328 92 + 1 UUGCGACUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGCGGCUGUUGCUUUUGAGCACGUUUACCUGUAAUGGCAUGCCACCAGCAGCGG--------- ...((.((((....(((.((((.((((.....))))..))))((((((((...((((....))))...)))))))).))).....)))))).--------- ( -24.70) >DroGri_CAF1 152251 87 + 1 CUGCUGUUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGUAACCGUUGCUCGCGAGCACGUUUACCUGCAAUGGCA-----GCAGCAGUAG--------- ((((((((((..(((((..((..((((.....)))))).((((.(((.(((....)))))).)))).)))))..)))-----)))))))...--------- ( -33.00) >DroEre_CAF1 142633 101 + 1 UUUUGACUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGCGGCUGUUGCUUUUGAGCACGUUUACCUGUAAUGGCAUGCCACCAGCAACAAAAACAACAA .......(((....))).((((.((((.....))))..))))..(((((((..((.(((.(((..........))).)))))..))))))).......... ( -21.60) >DroPer_CAF1 166787 92 + 1 GUGCGACUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGCGGCUGUUGCUUUUGAGCACGUUUACCUGUAAUGGCAUGCCACCAGCAGCAG--------- ((((...(((((.....))))).........(((((((.((....)).))))))).))))......((((..(((....)))...))))...--------- ( -24.90) >consensus UUGCGACUGCAUUUGCAAUGCAAUUUCAACACGAAAGCUGCGGCUGUUGCUUUUGAGCACGUUUACCUGUAAUGGCAUGCCACCAGCAGCAG_________ ............((((..((((.((((.....))))..))))((((((((...((((....))))...)))))))).........))))............ (-17.90 = -17.77 + -0.14)

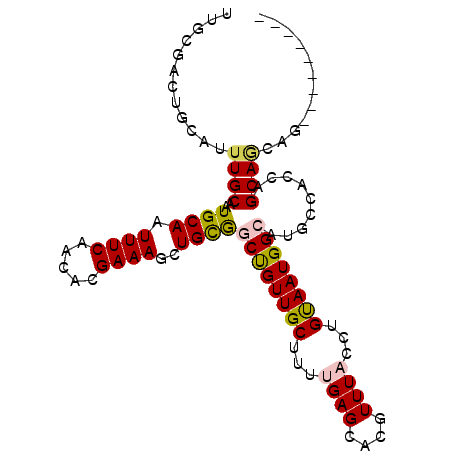

| Location | 18,107,039 – 18,107,131 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
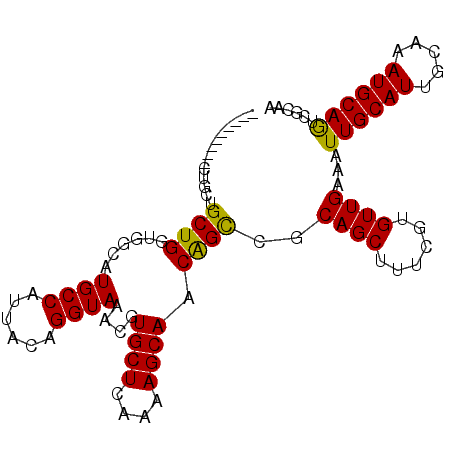
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -18.97 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18107039 92 - 27905053 ---------UUGUUGCUGGUGGCAUGCCAUUACAGGUAAACGUGCUCAAAAGCAACAGCCGCAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAGUCAAAA ---------((((((((..(((((((((......)))....)))).))..))))))))..((((.((((.....))))))))(((((....)))))..... ( -27.70) >DroVir_CAF1 151995 83 - 1 -------------UGCUGC-----UGCCAUUGCAGGUAAACGUGCUCGGCAGCAACGGUUGCAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAACGGCAG -------------((((((-----((((...(((.(....).)))..))))))....((((((......((((.......))))......)))))))))). ( -31.30) >DroPse_CAF1 141328 92 - 1 ---------CCGCUGCUGGUGGCAUGCCAUUACAGGUAAACGUGCUCAAAAGCAACAGCCGCAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAGUCGCAA ---------..(((((((((((....))))))..(((.....((((....))))...)))))))).............(((((((((....))))).)))) ( -29.10) >DroGri_CAF1 152251 87 - 1 ---------CUACUGCUGC-----UGCCAUUGCAGGUAAACGUGCUCGCGAGCAACGGUUACAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAACAGCAG ---------...((((((.-----(((..((((((((((.((((((....))).)))....((((......))))...)))).))))))..))).)))))) ( -31.00) >DroEre_CAF1 142633 101 - 1 UUGUUGUUUUUGUUGCUGGUGGCAUGCCAUUACAGGUAAACGUGCUCAAAAGCAACAGCCGCAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAGUCAAAA ..(((((..((((((((..(((((((((......)))....)))).))..))))))))..))))).......((((...(((((.....))))).)))).. ( -31.40) >DroPer_CAF1 166787 92 - 1 ---------CUGCUGCUGGUGGCAUGCCAUUACAGGUAAACGUGCUCAAAAGCAACAGCCGCAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAGUCGCAC ---------..(((((((((((....))))))..(((.....((((....))))...))))))))....((((....(((((((.....))))))).)))) ( -28.40) >consensus _________CUGCUGCUGGUGGCAUGCCAUUACAGGUAAACGUGCUCAAAAGCAACAGCCGCAGCUUUCGUGUUGAAAUUGCAUUGCAAAUGCAGUCGCAA ..............((((......((((......))))....((((....)))).))))..((((......))))...((((((.....))))))...... (-18.97 = -18.30 + -0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:13 2006