
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,059,662 – 18,059,766 |
| Length | 104 |
| Max. P | 0.785776 |

| Location | 18,059,662 – 18,059,766 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
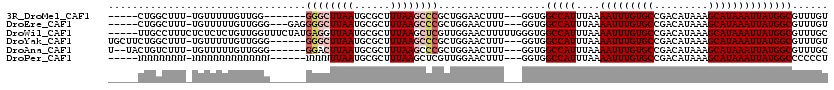
>3R_DroMel_CAF1 18059662 104 + 27905053 -----CUGGCUUU-UGUUUUUGUUGG-------GGGCUUAAUGCGCUUUAAGCCCGCUGGAACUUU---GGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCGUUUGU -----...(((..-.(((((....(.-------((((((((......)))))))).).)))))...---))).(((((....(((((((((.........))))))))))))))...... ( -32.10) >DroEre_CAF1 105304 108 + 1 -----CUGGCUUU-UGUUUUUGUUGGG---GAGGGGCUUAAUGCGCUUUAAGCCCGCUGGAACUUU---GGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCGUUUGU -----(..((.((-(.((......)).---)))((((((((......))))))))))..)......---....(((((....(((((((((.........))))))))))))))...... ( -30.20) >DroWil_CAF1 110792 115 + 1 -----UUGCCUUUCUCUCUCUGUUGGUUUCUAUGAGGUUAAUGCGCUUUAAGCUCGUUGGAACUUUUUGGGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCGUUUGC -----..........(.(((....((((((.(((((.((((......)))).))))).))))))....))).)(((((....(((((((((.........))))))))))))))...... ( -31.00) >DroYak_CAF1 106314 110 + 1 UGCUUCUGGCUUU-UGUUUUUGUUGGG------GGGCUUAAUGCGCUUUAAGCCCGCUGGAACUUU---GGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCGUUUGU .(((..(((((..-........(..(.------((((((((......)))))))).)..)......---...))))).....(((((((((.........)))))))))..)))...... ( -33.11) >DroAna_CAF1 98935 108 + 1 U--UACUGUCUUU-UGUUUUUGUUGGG------GGACUUAAUGCGCUUUAAGCCCGCUGGAACUUU---GGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCGUUUGC .--(((((.....-.(((((....(.(------((.(((((......)))))))).).)))))..)---))))(((((....(((((((((.........))))))))))))))...... ( -26.30) >DroPer_CAF1 125150 105 + 1 -----NNNNNNNN-NNNNNNNNNNNNN------NNNNUUAAUGCGCUUUAAGCUCGUUGGAACUUU---GGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCCCCCCU -----........-.............------....((((((.((.....)).))))))......---((.((((((....(((((((((.........)))))))))))))))))... ( -23.50) >consensus _____CUGGCUUU_UGUUUUUGUUGGG______GGGCUUAAUGCGCUUUAAGCCCGCUGGAACUUU___GGUGGCCAUUUAAAAUUUGUGCCGACAUAAAGCAUAAAUUAUGGCGUUUGU .................................((((((((......))))))))..................(((((....(((((((((.........))))))))))))))...... (-20.20 = -20.98 + 0.78)

| Location | 18,059,662 – 18,059,766 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
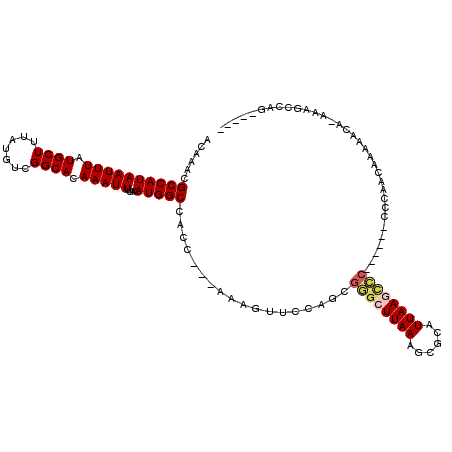
>3R_DroMel_CAF1 18059662 104 - 27905053 ACAAACGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACC---AAAGUUCCAGCGGGCUUAAAGCGCAUUAAGCCC-------CCAACAAAAACA-AAAGCCAG----- ......((((((((((.((((.......)))).)))))....)))))....---.........(.((((((((......))))))))-------.)..........-........----- ( -21.80) >DroEre_CAF1 105304 108 - 1 ACAAACGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACC---AAAGUUCCAGCGGGCUUAAAGCGCAUUAAGCCCCUC---CCCAACAAAAACA-AAAGCCAG----- ......((((((((((.((((.......)))).)))))....)))))....---...(((..((.((((((((......)))))))))).---...))).......-........----- ( -22.60) >DroWil_CAF1 110792 115 - 1 GCAAACGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACCCAAAAAGUUCCAACGAGCUUAAAGCGCAUUAACCUCAUAGAAACCAACAGAGAGAGAAAGGCAA----- ......(((.........(((((.....(((.((.........))))).......((((((....)))))))))))........(((.............))).......)))..----- ( -19.92) >DroYak_CAF1 106314 110 - 1 ACAAACGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACC---AAAGUUCCAGCGGGCUUAAAGCGCAUUAAGCCC------CCCAACAAAAACA-AAAGCCAGAAGCA ......((((((((((.((((.......)))).)))))....)))))....---...(((...(.((((((((......))))))))------)..))).......-...((.....)). ( -22.60) >DroAna_CAF1 98935 108 - 1 GCAAACGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACC---AAAGUUCCAGCGGGCUUAAAGCGCAUUAAGUCC------CCCAACAAAAACA-AAAGACAGUA--A ......((((((((((.((((.......)))).)))))....)))))....---...(((...(.((((((((......))))))))------)..))).......-..........--. ( -19.20) >DroPer_CAF1 125150 105 - 1 AGGGGGGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACC---AAAGUUCCAACGAGCUUAAAGCGCAUUAANNNN------NNNNNNNNNNNNN-NNNNNNNN----- .((..(((((((((((.((((.......)))).)))))....)))))).))---.((((((....))))))................------.............-........----- ( -21.60) >consensus ACAAACGCCAUAAUUUAUGCUUUAUGUCGGCACAAAUUUUAAAUGGCCACC___AAAGUUCCAGCGGGCUUAAAGCGCAUUAAGCCC______CCCAACAAAAACA_AAAGCCAG_____ ......((((((((((.((((.......)))).)))))....)))))..................((((((((......))))))))................................. (-16.70 = -17.20 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:01 2006