| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,032,971 – 18,033,073 |
| Length | 102 |
| Max. P | 0.901202 |

| Location | 18,032,971 – 18,033,073 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -26.65 |
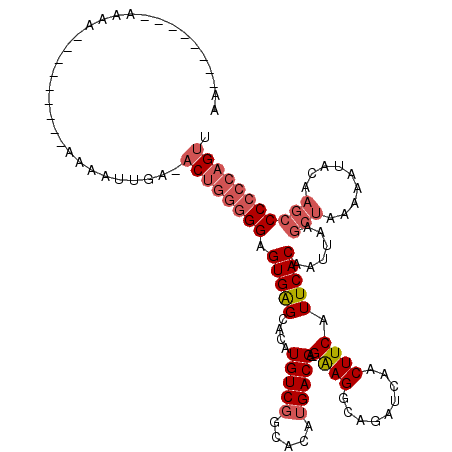
| Consensus MFE | -20.76 |
| Energy contribution | -21.90 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18032971 102 + 27905053 AAU-------AAAA---------AAAAUUGA-ACUUGGGGAGUGAGCACAUGUCGGCACAUGACGA-GAAGGCAGAUCAACUUCAUUCACAAUUAAGCUAAAAAUACAAGCCCCACAGUU ...-------....---------.......(-((((((((..(((((.(((((....)))))....-((((.........))))............))).......))..))))).)))) ( -21.41) >DroSec_CAF1 75540 95 + 1 -----------------------AAAAUUGA-ACUGGGGGAGUGAGCACAUGUCGGCACAUGACGA-GAAGGCAGAUCAACUUCAUUCACAAUUAAUCUAAAAAUACAAACCCCCCAGUU -----------------------.......(-((((((((.(((((..(((((....)))))....-((((.........)))).))))).(((........)))......))))))))) ( -26.40) >DroSim_CAF1 77461 95 + 1 -----------------------AAAAUUGA-ACUGGGGGAGUGAGCACAUGUCGGCACAUGACGA-GAAGGCAGAUCAACUUCAUUCACAAUUAAGCUAAAAAUACAAGCCCCCCAGUU -----------------------.......(-((((((((.(((((..(((((....)))))....-((((.........)))).)))))......(((.........)))))))))))) ( -29.60) >DroEre_CAF1 77436 119 + 1 AAAAACCGCCAAAAAAAAAAGAAAAAAUUGAAACUGGGGGAGUGGGCACAUGUCGGCACAUGACGA-GAAGGCAGAUCAACUUCAUUCACAAUUAAGCUAAAAAUACAAGCCCCCCAGUU ...............................(((((((((.(((((..(((((....)))))....-((((.........)))).)))))......(((.........)))))))))))) ( -28.50) >DroYak_CAF1 79264 110 + 1 UAAAGCCGCCAAAAA---------AAAUUGAAACUGGGGGAGUGAGCUCAUGUCGGCACAUGACGA-GAAGGCAGAUCAACUUCAUUCACAAUUAAGCUAAAAAUACAUGCCCCCCAGUU ...............---------.......(((((((((.(((((.((((((....))))))...-((((.........)))).)))))......((...........))))))))))) ( -29.60) >DroAna_CAF1 72091 103 + 1 AAU-------AAGA---------AAUAUUGCGACAGGGGGUGUGGGCUCAUGUCAGCACUCGACGGCGGAGGCAGAUCAACUUCAUUCACAAUUAAGCUAAAAAUACAAGCCC-CCAGCU ...-------....---------......((....((((((.(((.(((.((((.......))))...))).).((......))......................)).))))-)).)). ( -24.40) >consensus AA________AAAA_________AAAAUUGA_ACUGGGGGAGUGAGCACAUGUCGGCACAUGACGA_GAAGGCAGAUCAACUUCAUUCACAAUUAAGCUAAAAAUACAAGCCCCCCAGUU ................................((((((((.(((((....(((((.....)))))..((((.........)))).)))))......(((.........))))))))))). (-20.76 = -21.90 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:48 2006