
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,032,455 – 18,032,607 |
| Length | 152 |
| Max. P | 0.742090 |

| Location | 18,032,455 – 18,032,551 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18032455 96 + 27905053 CGUUGGCUUCAUCCAGCUUGAAUGCUUUCUCUGGCUGGUAAGUUCUGGCCAAGUUCUGGCGAAGGAACAAAACUAUUUCCUAAGCUGCUUAGCAAA .....(((.....((((((...((((..((.((((..(......)..))))))....)))).(((((.........)))))))))))...)))... ( -26.80) >DroSec_CAF1 75047 96 + 1 CGUUGGCUUCAUCCAGCUUGAAUGCUUUCUCUGGCUGGUUAGUUGUGGCCAAGUUCUGGCGCAGGAACAAAACUAUUUCCUAAGCUGCUUAGCAAA ..((((((.((.((((((.(((....)))...)))))).....)).))))))(((..((((((((((.........)))))..)).))).)))... ( -28.70) >DroSim_CAF1 76968 96 + 1 CGUUGGCUUCAUCCAGCUUGAAUGCUUUCUCUGGCUGGUUAGUUCUGGCCAAGUUCUGGCGCAGGAACAAUACUAUUUCCUAAGCUGCUUAGCAAA ..((((((....((((((.(((....)))...))))))........))))))(((..((((((((((.........)))))..)).))).)))... ( -27.00) >DroEre_CAF1 76945 93 + 1 CGUUGGCUUCAUCCAGCUCGAAUGCUUU---UGGCUGGUUAGUUCUGGCCAAGUUCUGGCGAAGGAACGUAACUAUUUCCUAAGCUGCUUAGACAA ..((((((....(((((.((((....))---)))))))........))))))(((..((((.(((((.........)))))....))))..))).. ( -26.40) >DroYak_CAF1 78749 96 + 1 CGUUGGCUUCAUCCAGCUUGAAUGCUUUCUCCGACUGGUUAGUUCUGGCCAAGUUCUGGUGAAGGAACGUAACUAUUUCCUAAGCUGCUUAGCCAA ..((((((.....(((((((......(((.(((((((((((....))))).))))..)).)))((((.........)))))))))))...)))))) ( -28.30) >consensus CGUUGGCUUCAUCCAGCUUGAAUGCUUUCUCUGGCUGGUUAGUUCUGGCCAAGUUCUGGCGAAGGAACAAAACUAUUUCCUAAGCUGCUUAGCAAA ..((((((....((((((.(((....)))...))))))........)))))).....(((..(((((.........)))))..))).......... (-22.76 = -23.00 + 0.24)

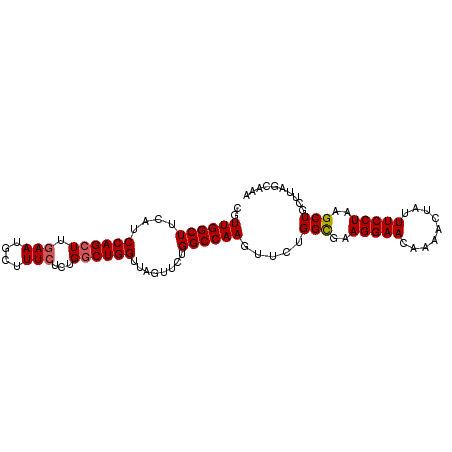

| Location | 18,032,517 – 18,032,607 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -13.53 |
| Energy contribution | -14.62 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18032517 90 + 27905053 AGGAACAAAACUAUUUCCUAAGCUGCUUAGCAAAAUGUCUAAUGCCUUGCUUAGACAAUUGCUUAAAGGAA---GAACUAGCGUAGUGAAAU----U (((((.........)))))..(((((..(((((..(((((((.((...))))))))).)))))...((...---...))...))))).....----. ( -19.20) >DroSec_CAF1 75109 93 + 1 AGGAACAAAACUAUUUCCUAAGCUGCUUAGCAAAAUGUCUAAUGCCUUGCUUAGACAAUUGCUAAAAGGAAGAAGAACUAGCGUAGAGAAAU----U ............((((((((.(((..(((((((..(((((((.((...))))))))).))))))).((.........))))).))).)))))----. ( -21.00) >DroSim_CAF1 77030 93 + 1 AGGAACAAUACUAUUUCCUAAGCUGCUUAGCAAAAUGUCUAAUGCCUUGCUUAGACAAUUGCUAAAAGGAAGAAGAACUAGCGUAGAGAAAU----U ............((((((((.(((..(((((((..(((((((.((...))))))))).))))))).((.........))))).))).)))))----. ( -21.00) >DroEre_CAF1 77004 90 + 1 AGGAACGUAACUAUUUCCUAAGCUGCUUAGACAAAUGUCUAAUGCCUUGCUUAGACAAUUGCUAAAAGGAA---GAUUUAGCGCCGAGUAAU----A .((..............((((((.((((((((....)))))).))...))))))......((((((.....---..)))))).)).......----. ( -22.60) >DroYak_CAF1 78811 89 + 1 AGGAACGUAACUAUUUCCUAAGCUGCUUAGCCAAAUGUCUAAUGCUUUGCUUAGACAAUUGCUAAAAGGAA---GAUUUAGAA-AGAGAAAU----U ..........(((..((.....((..(((((.((.(((((((.((...))))))))).)))))))..))..---))..)))..-........----. ( -16.90) >DroAna_CAF1 71691 89 + 1 AGGAA-----CUAUUUCCUAAGCUGCUUACGGAGCUGUGUAAUGCCCUGCUUAGACAAUUGUUAAAGGGAGAA-AAUUCA--AAAGGGAAACUAACU (((((-----....))))).((((.(....).))))((......((((.(((.(((....))).)))(((...-..))).--..))))......)). ( -17.70) >consensus AGGAACAAAACUAUUUCCUAAGCUGCUUAGCAAAAUGUCUAAUGCCUUGCUUAGACAAUUGCUAAAAGGAA___GAACUAGCGUAGAGAAAU____U (((((.........)))))...((..(((((((..(((((((.((...))))))))).)))))))..))............................ (-13.53 = -14.62 + 1.08)


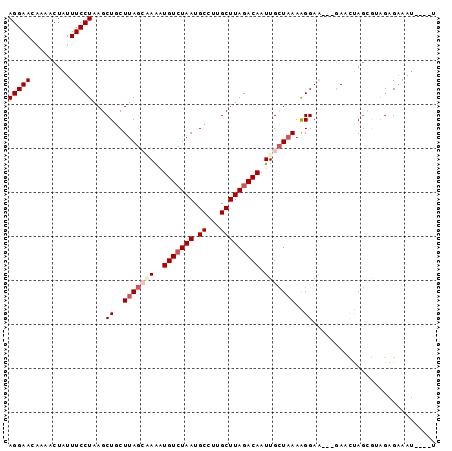
| Location | 18,032,517 – 18,032,607 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18032517 90 - 27905053 A----AUUUCACUACGCUAGUUC---UUCCUUUAAGCAAUUGUCUAAGCAAGGCAUUAGACAUUUUGCUAAGCAGCUUAGGAAAUAGUUUUGUUCCU .----(((((.(((.(((.(((.---........(((((.(((((((((...)).)))))))..))))).)))))).))))))))............ ( -20.40) >DroSec_CAF1 75109 93 - 1 A----AUUUCUCUACGCUAGUUCUUCUUCCUUUUAGCAAUUGUCUAAGCAAGGCAUUAGACAUUUUGCUAAGCAGCUUAGGAAAUAGUUUUGUUCCU .----(((((.(((.(((.((...........(((((((.(((((((((...)).)))))))..)))))))))))).))))))))............ ( -22.90) >DroSim_CAF1 77030 93 - 1 A----AUUUCUCUACGCUAGUUCUUCUUCCUUUUAGCAAUUGUCUAAGCAAGGCAUUAGACAUUUUGCUAAGCAGCUUAGGAAAUAGUAUUGUUCCU .----(((((.(((.(((.((...........(((((((.(((((((((...)).)))))))..)))))))))))).))))))))............ ( -22.90) >DroEre_CAF1 77004 90 - 1 U----AUUACUCGGCGCUAAAUC---UUCCUUUUAGCAAUUGUCUAAGCAAGGCAUUAGACAUUUGUCUAAGCAGCUUAGGAAAUAGUUACGUUCCU .----.......(((((((((..---.....)))))).(((..((((((...((.((((((....)))))))).))))))..))).)))........ ( -24.00) >DroYak_CAF1 78811 89 - 1 A----AUUUCUCU-UUCUAAAUC---UUCCUUUUAGCAAUUGUCUAAGCAAAGCAUUAGACAUUUGGCUAAGCAGCUUAGGAAAUAGUUACGUUCCU .----........-.........---(((((((((((...(((((((((...)).)))))))....))))))......))))).............. ( -17.40) >DroAna_CAF1 71691 89 - 1 AGUUAGUUUCCCUUU--UGAAUU-UUCUCCCUUUAACAAUUGUCUAAGCAGGGCAUUACACAGCUCCGUAAGCAGCUUAGGAAAUAG-----UUCCU .....((((((....--......-................(((((.....)))))......((((.(....).))))..))))))..-----..... ( -14.20) >consensus A____AUUUCUCUACGCUAAAUC___UUCCUUUUAGCAAUUGUCUAAGCAAGGCAUUAGACAUUUUGCUAAGCAGCUUAGGAAAUAGUUUUGUUCCU .....(((((.....(((((............)))))......((((((...((.((((........)))))).)))))))))))............ (-13.84 = -13.90 + 0.06)

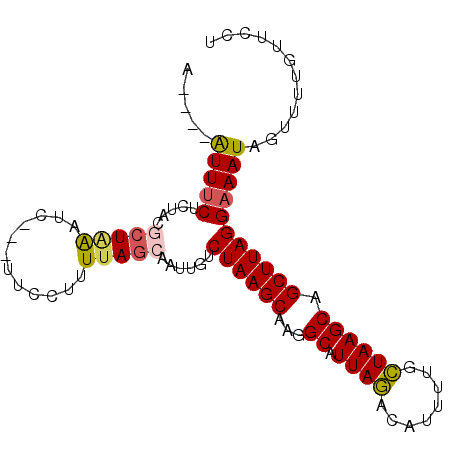
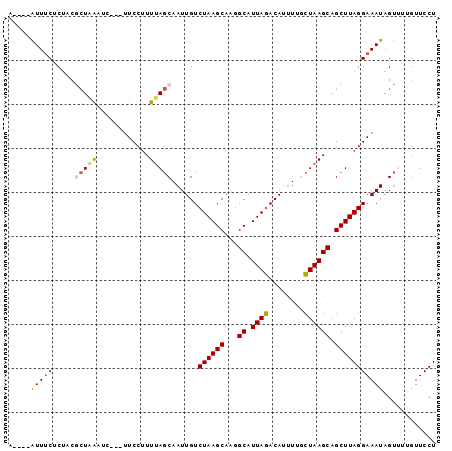
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:47 2006