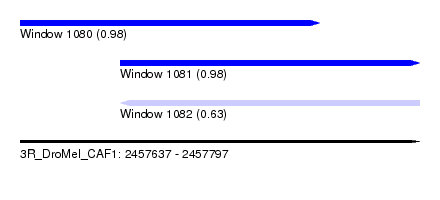
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,457,637 – 2,457,797 |
| Length | 160 |
| Max. P | 0.982062 |

| Location | 2,457,637 – 2,457,757 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -36.88 |
| Energy contribution | -38.45 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2457637 120 + 27905053 UAACCUCGAAAUCGAGAGCGCAGAACUGAAAGUUGGCCAUCAAGUUGUGUGUGGGGCAAUCAGAGAGAUGCCAGAAAUCGGAUUGCUUGGCACAACAAUUUGGUGACCCAGAGUCAUUGA ....((((....))))..........(((...((((.(((((((((((((((.((((((((.....(((.......))).)))))))).)))).)))))))))))..))))..))).... ( -42.20) >DroSec_CAF1 16647 120 + 1 UAACCUCGAAAUCGAGAGCGCAGAACUGAAAGUUGGCCAUCAUGUUGUGUGUGGGGCAAUCAGAGAAAUGCCAGAAAUCGGAUUGCCUGGCACAACAAUUUGGGGACCCAGAGUCAUUGA ....((((....))))..........(((...((((((.(((.(((((((((.((((((((.((.............)).)))))))).)))).))))).)))))..))))..))).... ( -38.42) >DroSim_CAF1 13372 120 + 1 UAACCUCGAAAUCGAGAGCGCAGAACUGAAAGUUGGCCAUCAUGUUGUGUGUGGGGCAAUCAGAGAAAUGCCAGAAAUCGGAUUGCCUGGCACAACAAUUUGGUGACCCAGAGUCAUUGA ....((((....))))..........(((...((((.(((((.(((((((((.((((((((.((.............)).)))))))).)))).))))).)))))..))))..))).... ( -40.92) >DroEre_CAF1 18139 117 + 1 CAACCUCGAAAUCGAGAGCGCAGAACU-AAAGUUGGCCAUCAAGUUGU--GUGGGGCAAUCAGAGAGAUGCCAGAAAUCGGAUUGCCUGGAACAACAAUUUGGUGACCCAGAGUCAUUGA ....((((....))))...........-....((((.(((((((((((--.(.((((((((.....(((.......))).)))))))).)....)))))))))))..))))......... ( -34.70) >consensus UAACCUCGAAAUCGAGAGCGCAGAACUGAAAGUUGGCCAUCAAGUUGUGUGUGGGGCAAUCAGAGAAAUGCCAGAAAUCGGAUUGCCUGGCACAACAAUUUGGUGACCCAGAGUCAUUGA ....((((....))))..........(((...((((.(((((((((((((((.((((((((.((.............)).)))))))).)))).)))))))))))..))))..))).... (-36.88 = -38.45 + 1.56)


| Location | 2,457,677 – 2,457,797 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -47.66 |
| Consensus MFE | -45.18 |
| Energy contribution | -45.74 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2457677 120 + 27905053 CAAGUUGUGUGUGGGGCAAUCAGAGAGAUGCCAGAAAUCGGAUUGCUUGGCACAACAAUUUGGUGACCCAGAGUCAUUGACACGCCUCGUUUUCGGGGCAUUGGCUGGGCAUUAAAUUCA ((((((((((((.((((((((.....(((.......))).)))))))).)))).))))))))(((.(((((.(((...)))..((((((....)))))).....))))))))........ ( -48.70) >DroSec_CAF1 16687 120 + 1 CAUGUUGUGUGUGGGGCAAUCAGAGAAAUGCCAGAAAUCGGAUUGCCUGGCACAACAAUUUGGGGACCCAGAGUCAUUGACACGCCUCGUUUUCGGGGCAUUGGCUGGGCAUUAAAUUCA ((.(((((((((.((((((((.((.............)).)))))))).)))).))))).))..(.(((((.(((...)))..((((((....)))))).....)))))).......... ( -45.62) >DroSim_CAF1 13412 120 + 1 CAUGUUGUGUGUGGGGCAAUCAGAGAAAUGCCAGAAAUCGGAUUGCCUGGCACAACAAUUUGGUGACCCAGAGUCAUUGACACGCCUCGUUGUCGGGGCAUUGGCUGGGCAUUAAAUUCA .......(((((.((((((((.((.............)).)))))))).)))))..(((((((((.(((((.(((...)))..((((((....)))))).....)))))))))))))).. ( -48.62) >DroEre_CAF1 18178 118 + 1 CAAGUUGU--GUGGGGCAAUCAGAGAGAUGCCAGAAAUCGGAUUGCCUGGAACAACAAUUUGGUGACCCAGAGUCAUUGACACGCCUCGUUUUCGGGGCAUUGGCUGGGCAUUAAAUUCA ...(((((--.(.((((((((.....(((.......))).)))))))).).)))))(((((((((.(((((.(((...)))..((((((....)))))).....)))))))))))))).. ( -47.70) >consensus CAAGUUGUGUGUGGGGCAAUCAGAGAAAUGCCAGAAAUCGGAUUGCCUGGCACAACAAUUUGGUGACCCAGAGUCAUUGACACGCCUCGUUUUCGGGGCAUUGGCUGGGCAUUAAAUUCA ...(((((((...((((((((.((.............)).)))))))).)))))))(((((((((.(((((.(((...)))..((((((....)))))).....)))))))))))))).. (-45.18 = -45.74 + 0.56)

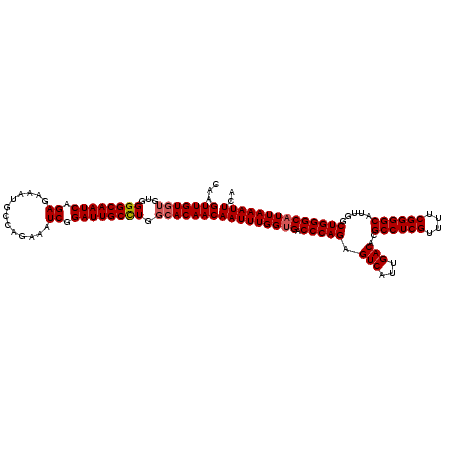
| Location | 2,457,677 – 2,457,797 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -31.27 |
| Energy contribution | -31.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2457677 120 - 27905053 UGAAUUUAAUGCCCAGCCAAUGCCCCGAAAACGAGGCGUGUCAAUGACUCUGGGUCACCAAAUUGUUGUGCCAAGCAAUCCGAUUUCUGGCAUCUCUCUGAUUGCCCCACACACAACUUG ..(((((..(((((((...(((((.((....)).)))))(((...))).))))).))..)))))((((((....((((((.((.............)).))))))......))))))... ( -30.62) >DroSec_CAF1 16687 120 - 1 UGAAUUUAAUGCCCAGCCAAUGCCCCGAAAACGAGGCGUGUCAAUGACUCUGGGUCCCCAAAUUGUUGUGCCAGGCAAUCCGAUUUCUGGCAUUUCUCUGAUUGCCCCACACACAACAUG ..........((((((...(((((.((....)).)))))(((...))).))))))........(((((((...(((((((.((.............)).))))))).....))))))).. ( -34.02) >DroSim_CAF1 13412 120 - 1 UGAAUUUAAUGCCCAGCCAAUGCCCCGACAACGAGGCGUGUCAAUGACUCUGGGUCACCAAAUUGUUGUGCCAGGCAAUCCGAUUUCUGGCAUUUCUCUGAUUGCCCCACACACAACAUG ..(((((..(((((((...(((((.((....)).)))))(((...))).))))).))..)))))((((((...(((((((.((.............)).))))))).....))))))... ( -34.62) >DroEre_CAF1 18178 118 - 1 UGAAUUUAAUGCCCAGCCAAUGCCCCGAAAACGAGGCGUGUCAAUGACUCUGGGUCACCAAAUUGUUGUUCCAGGCAAUCCGAUUUCUGGCAUCUCUCUGAUUGCCCCAC--ACAACUUG ..(((((..(((((((...(((((.((....)).)))))(((...))).))))).))..)))))(((((....(((((((.((.............)).)))))))....--)))))... ( -32.32) >consensus UGAAUUUAAUGCCCAGCCAAUGCCCCGAAAACGAGGCGUGUCAAUGACUCUGGGUCACCAAAUUGUUGUGCCAGGCAAUCCGAUUUCUGGCAUCUCUCUGAUUGCCCCACACACAACAUG ..(((((...((((((...(((((.((....)).)))))(((...))).))))))....)))))((((((...(((((((.((.............)).))))))).....))))))... (-31.27 = -31.77 + 0.50)

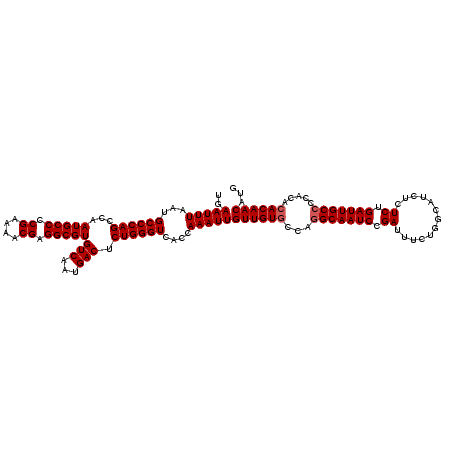
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:15 2006