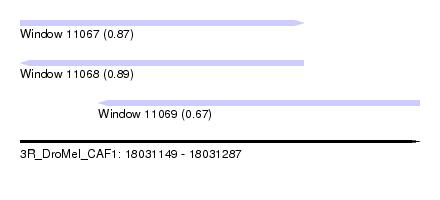
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,031,149 – 18,031,287 |
| Length | 138 |
| Max. P | 0.892178 |

| Location | 18,031,149 – 18,031,247 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.32 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
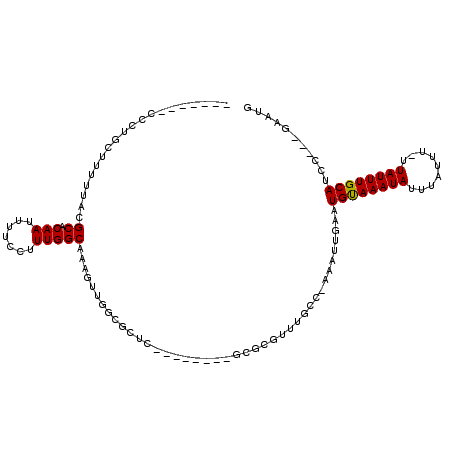
>3R_DroMel_CAF1 18031149 98 + 27905053 AGAUACUAACGCACAC------------UCGCAUGCAUGCAGUCGGAUGCAAAUAAAAAUAAAUAUUUACAUUCAAUUUCGACAAACGCGCCAGCCACUGAGCGCCAACU ..........(((..(------------......)..))).((((((((.(((((........))))).))))......))))....(((((((...))).))))..... ( -17.10) >DroSec_CAF1 73793 94 + 1 AGAUACUGACGCACAC------------UCGCA----UGCAGUCGGAUGCAAAUAAAAAUAAAUAUUUACAUUCAAUUUCGACAAACGCGCCAGCCACUGAGCGCCAACU .......((((((...------------.....----))).)))(((((.(((((........))))).))))).............(((((((...))).))))..... ( -19.00) >DroSim_CAF1 75698 94 + 1 AGAUACUAACGCACAC------------UCGCA----UGCAGUCGGAUGCAAAUAAAAAUAAAUAUUUACAUUCAAUUUCGACAAACGCGCCAGCCACUGAGCGCCAACU ..........(((...------------.....----))).((((((((.(((((........))))).))))......))))....(((((((...))).))))..... ( -16.60) >DroEre_CAF1 75607 94 + 1 AGAUACUAACGCAUAC------------UCGCA----UGCAGUCGAAUGCAAAUAAAAAUAAAUAUUUACAUUCAAUUUCGACAAACGCGCCAGCCACUGAGCGCCAACU ..........((((..------------....)----))).((((((((.(((((........))))).)))......)))))....(((((((...))).))))..... ( -18.70) >DroYak_CAF1 77376 106 + 1 AGAUACUAACGCAUACUCUCGCACUCACUCGCA----UGCAGUCGAAUGCAAAUAAAAAUAAAUAUUUACAUUCAAUUUCGACAAACGCGGCAGCCACUGAGCGCCAACU ....................((.((((...((.----(((....(((((.(((((........))))).))))).....((.(....))))))))...)))).))..... ( -18.70) >consensus AGAUACUAACGCACAC____________UCGCA____UGCAGUCGGAUGCAAAUAAAAAUAAAUAUUUACAUUCAAUUUCGACAAACGCGCCAGCCACUGAGCGCCAACU ..............................(((....))).((((((((.(((((........))))).))......))))))....(((((((...))).))))..... (-15.36 = -15.32 + -0.04)



| Location | 18,031,149 – 18,031,247 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.28 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18031149 98 - 27905053 AGUUGGCGCUCAGUGGCUGGCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUUUUAUUUGCAUCCGACUGCAUGCAUGCGA------------GUGUGCGUUAGUAUCU ..(((((((.((.(.((..(((.((..((((.....((.((((((((........)))))))))))))).)).)))..)).)------------.)).)))))))..... ( -28.80) >DroSec_CAF1 73793 94 - 1 AGUUGGCGCUCAGUGGCUGGCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUUUUAUUUGCAUCCGACUGCA----UGCGA------------GUGUGCGUCAGUAUCU ..(((((((.((.(.((..((.((.....))...(((.(((((((((........))))))))).)))..)).----.)).)------------.)).)))))))..... ( -27.10) >DroSim_CAF1 75698 94 - 1 AGUUGGCGCUCAGUGGCUGGCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUUUUAUUUGCAUCCGACUGCA----UGCGA------------GUGUGCGUUAGUAUCU ..(((((((.((.(.((..((.((.....))...(((.(((((((((........))))))))).)))..)).----.)).)------------.)).)))))))..... ( -25.00) >DroEre_CAF1 75607 94 - 1 AGUUGGCGCUCAGUGGCUGGCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUUUUAUUUGCAUUCGACUGCA----UGCGA------------GUAUGCGUUAGUAUCU ...............((((((((((...(((...(((((((((((((........))))))))))))).....----..)))------------..)))))))))).... ( -27.90) >DroYak_CAF1 77376 106 - 1 AGUUGGCGCUCAGUGGCUGCCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUUUUAUUUGCAUUCGACUGCA----UGCGAGUGAGUGCGAGAGUAUGCGUUAGUAUCU ..((.(((((((.(.(((((..((.....))...(((((((((((((........)))))))))))))..)))----.)).).))))))).))((.((((....)))))) ( -34.90) >consensus AGUUGGCGCUCAGUGGCUGGCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUUUUAUUUGCAUCCGACUGCA____UGCGA____________GUGUGCGUUAGUAUCU ...............(((((((((...((((.....((.((((((((........))))))))))))))((((....))))................))))))))).... (-22.48 = -22.28 + -0.20)



| Location | 18,031,176 – 18,031,287 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -8.54 |
| Energy contribution | -8.40 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18031176 111 - 27905053 CUGUUCCCGCUGCUUUUUACGCACAAUUUUCCUUUGGCAAAGUUGGCGCUCAGUGGCUGGCGCGUUUGUCGAAAUUGAAUGUAAAUAUUUAUUU-UUAUUUGCAUCC---GACUG ..((.((.((..((.....(((.(((((((((...)).))))))))))...))..)).)).))....((((.....((.((((((((.......-.)))))))))))---))).. ( -29.40) >DroVir_CAF1 84763 89 - 1 -------CAACGCUUUUUACGCACAAUUUUCCUUUGGCAA-GCAG-C------------GCGCGUUUGCC-AAAUGGAAUGUAAAUAUUUAUUU-UUAUUUGCAGUC---AGCCG -------....(((..............((((((((((((-((..-.------------....)))))))-))).))))((((((((.......-.))))))))...---))).. ( -25.30) >DroPse_CAF1 95944 95 - 1 --------CCUCCUUUUUACGCACAAUUUUCCUUUGGCAAAGUUUGCGCUCC-------GCGCGUUUGCC-AAAUUGAAUGUAAAUAUUUAUUU-UUAUUUGCAUCC---GAAUG --------........................(((((((((...(((((...-------)))))))))))-)))(((.(((((((((.......-.))))))))).)---))... ( -24.00) >DroWil_CAF1 79590 88 - 1 -------CGAUGCUUUUUACGCACAAUUUUCCUUUGGCAA-GUUG--------------GCGCGUUUG-CGAAAUUGAAUGCAAAUAUUUAUUUUUUAUUUGCAGCC---GCA-A -------...(((.....((((.((((((.((...)).))-))))--------------..))))...-.......(..((((((((.........))))))))..)---)))-. ( -20.20) >DroAna_CAF1 70428 105 - 1 CCGCUCCCCCUGCUUUUUACGCACAAUUUUCCUUUGGCA-AGUUGGCGCUCGCUG--------GUUUGUCGAAAUUGAAUGUAAAUAUUUAUUU-UUAUUUGCAUCUGUUGGCUG ..((.......)).......((.((((.....(((((((-((..(((....))).--------.))))))))).....(((((((((.......-.)))))))))..)))))).. ( -26.00) >DroPer_CAF1 92319 98 - 1 -----CUGCCUCCUUUUUACGCACAAUUUUCCUUUGGCAAAGUUUGCGCUCC-------GCGCGUUUGCC-AAAUUGAAUGUAAAUAUUUAUUU-UUAUUUGCAUCC---GAAUG -----...........................(((((((((...(((((...-------)))))))))))-)))(((.(((((((((.......-.))))))))).)---))... ( -24.00) >consensus _______CCCUGCUUUUUACGCACAAUUUUCCUUUGGCAAAGUUGGCGCUC________GCGCGUUUGCC_AAAUUGAAUGUAAAUAUUUAUUU_UUAUUUGCAUCC___GAAUG ....................((.(((.......))))).........................................((((((((.........))))))))........... ( -8.54 = -8.40 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:43 2006