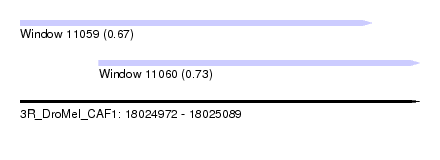
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,024,972 – 18,025,089 |
| Length | 117 |
| Max. P | 0.734804 |

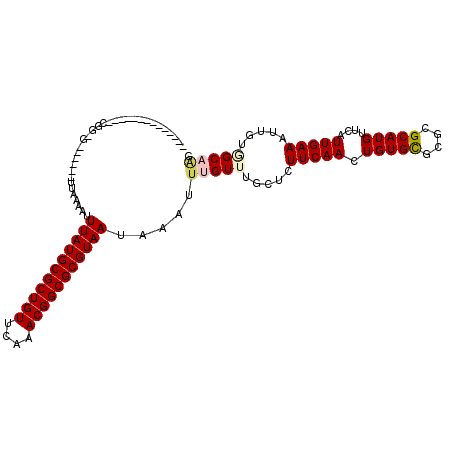
| Location | 18,024,972 – 18,025,075 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18024972 103 + 27905053 A---CCUCCACACAUGG-G------UUAAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGCGCAUGUUCAUUGAAAUUGUGGCAAG .---...(((((...((-(------(..(((((((((((((((....))))))))))...)))))....))))(((((..(..((....))..)...)))))..))))).... ( -28.50) >DroVir_CAF1 78092 92 + 1 -------------ACUG--------GUUAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGCGCGCGCAUGUUCAUUGAAUUUGUUGCAAG -------------....--------...........(((((((....)))))))((((((((((((..((......(((((((....)))).))))).))))))))))))... ( -23.60) >DroPse_CAF1 86977 94 + 1 --------------UGG-G----CCUUAAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGAGCAUGUUCAUUGAAAUUGUGGCAAC --------------...-(----((.(((...(((((((((((....)))))))))))..(((.(((((((.(.........).))))))).)))........))).)))... ( -27.80) >DroWil_CAF1 73756 109 + 1 UGUAGUGGACC---AGG-GAUAGCUCUUAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGCGCAUGUUCAUUGAAAUUGUUGCCGG ..((((((((.---..(-((.(((...((((((((((((((((....))))))))))...))))))...))).)))...((((....)))).))))))))............. ( -30.90) >DroMoj_CAF1 86214 92 + 1 -------------GCUGCU------GUUAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGCGCGCGCAUGUUCAUUGAAUUUGUAGCU-- -------------(((((.------.((((..(((((((((((....)))))))))))..................(((((((....)))).)))..))))....))))).-- ( -26.50) >DroPer_CAF1 85153 94 + 1 --------------GGG-G----CCUUAAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGCGCGAGCAUGUUCAUUGAAAUUGUGGCAAC --------------...-(----((.(((...(((((((((((....)))))))))))...............(((((.(((((....)))))....))))).))).)))... ( -27.80) >consensus ______________CGG_G______UUAAAAUUUAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGCGCGCGCAUGUUCAUUGAAAUUGUGGCAAG ................................(((((((((((....))))))))))).....(((((.....(((((.(((((....)))))....))))).....))))). (-20.46 = -20.85 + 0.39)

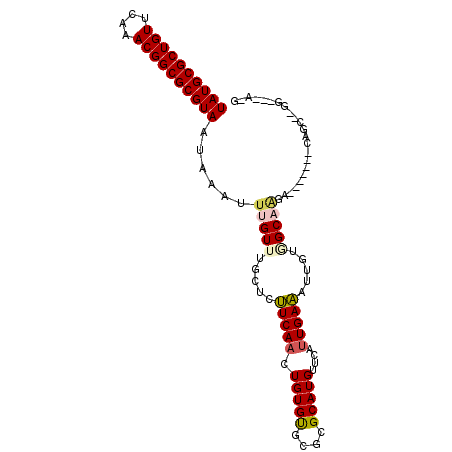
| Location | 18,024,995 – 18,025,089 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18024995 94 + 27905053 UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGCGCAUGUUCAUUGAAAUUGUGGCAAGA-------GAGG-UGGUGAAUG ((((((((((....))))))))))........(((..((((((.......(((.((((..(((...)))..)))))))...-------))))-.))..))). ( -27.80) >DroVir_CAF1 78104 85 + 1 UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGCGCGCGCAUGUUCAUUGAAUUUGUUGCAAGC-------CAGC---------- ((((((((((....))))))))))........((((((..(((((.(((((....)))))....)))))......))))))-------....---------- ( -24.60) >DroPse_CAF1 86991 95 + 1 UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGAGCAUGUUCAUUGAAAUUGUGGCAACA-------CACAGAGACACAGA ((((((((((....)))))))))).....(((((...((((((((..(..((....))..)...)))))..((((....))-------))..)))..))))) ( -30.00) >DroWil_CAF1 73785 100 + 1 UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGCGCAUGUUCAUUGAAAUUGUUGCCGGUUUGCUGUCAGC-AGGCAAA-G ((((((((((....)))))))))).........((((((.(((((..(..((....))..)...)))))..(((((.(((....))).))))-)))))))-. ( -31.40) >DroMoj_CAF1 86228 83 + 1 UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGCGCGCGCAUGUUCAUUGAAUUUGUAGCU--G-------CAGC---------- ((((((((((....)))))))))).....................(((((.((..(((.((((...)))).))).)))--)-------))).---------- ( -23.50) >DroAna_CAF1 64620 91 + 1 UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCCUCAACUGUGUGCGCGCAUGUUCACUGAGGUUGUGGCAGC----------GG-GGGUGGGCG ((((((((((....))))))))))........(((..((((((((((..(((.((....)).)))...))))).......----------))-)))..))). ( -33.81) >consensus UAUGCGCUGUUCAAACGGCGCGUAAUAAAUUUGUUUGCUCUUCAACUGUGUGCGCGCAUGUUCAUUGAAAUUGUGGCAAGA_______CAGC__GG___A_G ((((((((((....))))))))))......(((((.....(((((.(((((....)))))....))))).....)))))....................... (-19.05 = -19.30 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:34 2006