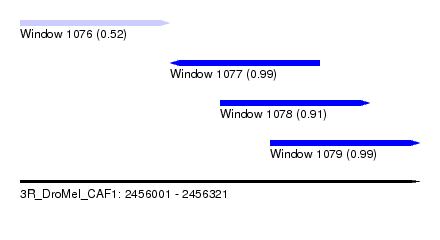
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,456,001 – 2,456,321 |
| Length | 320 |
| Max. P | 0.988293 |

| Location | 2,456,001 – 2,456,121 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2456001 120 + 27905053 CAACAACAAGUUCCUCACCAUGUACAGUACUUUGAUCAAGAGCUUCGCUGCCUCCACGGAGUUUUCCACCAACGAAGUGGUCAGCUUGCUGAUCGUGUUUUACAAGUACGCCCUGAACAA .........((((.......(((((...((((((....(((((((((.((....))))))))))).......))))))((((((....))))))...........)))))....)))).. ( -26.40) >DroSec_CAF1 14998 120 + 1 CAACAACAAGUUCCUCACCAUGUACAGUACUCUGAUCAAGAGCUUCGCUGCCUCCACGGAGUUUUCCACCAGCGAAGUGGUCAGCCUGCUGAUCGUGUUCUACAAGUACGCCCUGAACAA .........((((...........(((((..((((((....(((((((((.(((....)))........)))))))))))))))..)))))...((((.(.....).))))...)))).. ( -33.20) >DroSim_CAF1 11717 120 + 1 CAACAACAAGUUCCUCACCAUGUACAGUACUCUGAUCAAGAGCUUCGCUGCCUCCACGGAGUUUUCCACCAGCGAAGUGGUCAGCUUGCUGAUCGUGUUCUACAAGUACGCCCUGAACAA .........((((...........(((((..((((((....(((((((((.(((....)))........)))))))))))))))..)))))...((((.(.....).))))...)))).. ( -33.20) >DroEre_CAF1 15952 120 + 1 CAACAACAAGUUCCUCACCAUGUACAGUACUCUGAUCAAGAGCUUCGCUGCCUCCACGGAGUUUUCCACCAGCGAAGUGGUCAGCUUGCUGAUCGUGUUCUACAAAUACGCCCUCAACAA ....................(((.(((((..((((((....(((((((((.(((....)))........)))))))))))))))..)))))..((((((.....))))))......))). ( -31.20) >consensus CAACAACAAGUUCCUCACCAUGUACAGUACUCUGAUCAAGAGCUUCGCUGCCUCCACGGAGUUUUCCACCAGCGAAGUGGUCAGCUUGCUGAUCGUGUUCUACAAGUACGCCCUGAACAA .........((((...........(((((..((((((....(((((((((.(((....)))........)))))))))))))))..)))))...((((.........))))...)))).. (-29.46 = -29.78 + 0.31)

| Location | 2,456,121 – 2,456,241 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -48.57 |
| Consensus MFE | -47.67 |
| Energy contribution | -48.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2456121 120 - 27905053 ACUGAUCGGCCAUCCUGCGUGAUAUUCAUCGAAAUCCGGUCGAUGAUGGUCACGUCGAAGAUCCCAAAGCUGACGAGGAAUAGAUUGUACAGCUGGCUGGUGGACAUCAUGCGGGACCUG .......((...(((((((((((.(((((((....((((((..(((((....)))))..))))....(((((((((........)))).))))))).))))))).))))))))))))).. ( -44.00) >DroSec_CAF1 15118 120 - 1 ACUGAUCGGCCGUCCUGCGUGAUGUUCAUCGAAAUGCGGUCGAUGAUGGUCACGUCGAAGAUCCCAAAGCUGACGAGGAAUAGAUUGUACAGCUGGCUGGUGGACAUCAUGCGGGAUCUG ......(((..((((((((((((((((((((...((.((((..(((((....)))))..)))).)).(((((((((........)))).)))))...))))))))))))))))))))))) ( -51.10) >DroSim_CAF1 11837 120 - 1 ACUGAUCGGCCGUCCUGCGUGAUGUUCAUCGAAAUGCGGUCGAUGAUGGUCACGUCGAAGAUCCCAAAGCUGACGAGGAAUAGAUUGUACAGCUGGCUGGUGGACAUCAUGCGGGAUCUG ......(((..((((((((((((((((((((...((.((((..(((((....)))))..)))).)).(((((((((........)))).)))))...))))))))))))))))))))))) ( -51.10) >DroEre_CAF1 16072 120 - 1 ACUGAUCGGCCGUCCUGCGUGAUAUUCAUCGAAAUGCGGUCGAUGAUGGUGACGUCGAAGAUCCCAAAGCUGACGAGGAAUAGAUUGUACAGCUGACUGGUGGACAUCAUGCGGGAUCUG ......(((..((((((((((((.(((((((...((.((((..(((((....)))))..)))).)).(((((((((........)))).)))))...))))))).))))))))))))))) ( -48.10) >consensus ACUGAUCGGCCGUCCUGCGUGAUAUUCAUCGAAAUGCGGUCGAUGAUGGUCACGUCGAAGAUCCCAAAGCUGACGAGGAAUAGAUUGUACAGCUGGCUGGUGGACAUCAUGCGGGAUCUG ......(((..((((((((((((((((((((...((.((((..(((((....)))))..)))).)).(((((((((........)))).)))))...))))))))))))))))))))))) (-47.67 = -48.68 + 1.00)

| Location | 2,456,161 – 2,456,281 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -42.43 |
| Consensus MFE | -41.55 |
| Energy contribution | -41.43 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2456161 120 + 27905053 UUCCUCGUCAGCUUUGGGAUCUUCGACGUGACCAUCAUCGACCGGAUUUCGAUGAAUAUCACGCAGGAUGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUC ......((((.(.((((.....)))).)))))..(((((((.......)))))))....(((((((((((((((.........(((((....)))))..)))))))))))))))...... ( -40.20) >DroSec_CAF1 15158 120 + 1 UUCCUCGUCAGCUUUGGGAUCUUCGACGUGACCAUCAUCGACCGCAUUUCGAUGAACAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUC ......((((.(.((((.....)))).)))))..(((((((.......)))))))....(((((((((((((((.........(((((....)))))..)))))))))))))))...... ( -42.70) >DroSim_CAF1 11877 120 + 1 UUCCUCGUCAGCUUUGGGAUCUUCGACGUGACCAUCAUCGACCGCAUUUCGAUGAACAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUC ......((((.(.((((.....)))).)))))..(((((((.......)))))))....(((((((((((((((.........(((((....)))))..)))))))))))))))...... ( -42.70) >DroEre_CAF1 16112 120 + 1 UUCCUCGUCAGCUUUGGGAUCUUCGACGUCACCAUCAUCGACCGCAUUUCGAUGAAUAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGCGUGUUCUUC ..............((((((.......))).)))(((((((.......)))))))....(((((((((((((((.........(((((....)))))..)))))))))))))))...... ( -44.10) >consensus UUCCUCGUCAGCUUUGGGAUCUUCGACGUGACCAUCAUCGACCGCAUUUCGAUGAACAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUC ......((((.(.((((.....)))).)))))..(((((((.......)))))))....(((((((((((((((.........(((((....)))))..)))))))))))))))...... (-41.55 = -41.43 + -0.12)

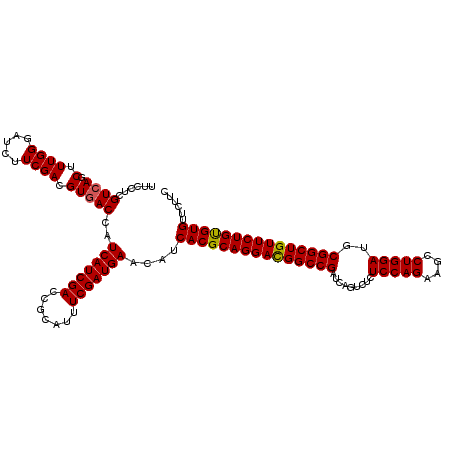
| Location | 2,456,201 – 2,456,321 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -50.30 |
| Consensus MFE | -48.60 |
| Energy contribution | -48.48 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2456201 120 + 27905053 ACCGGAUUUCGAUGAAUAUCACGCAGGAUGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUCAACGGAAGUCUGCAGGAGCGAAUGAAGUUCGCAUUUGAGG ..(((((((((.((((...(((((((((((((((.........(((((....)))))..)))))))))))))))...)))).).))))))))((((.((((((...)))))).))))... ( -48.60) >DroSec_CAF1 15198 120 + 1 ACCGCAUUUCGAUGAACAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUCAACGGAAGUCUGCAGGAGCGAAUGAAGUUCGCAUUUGAGG .((((((((((.((((...(((((((((((((((.........(((((....)))))..)))))))))))))))...)))).)))))...))).)).((((((...))))))........ ( -50.00) >DroSim_CAF1 11917 120 + 1 ACCGCAUUUCGAUGAACAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUCAACGGAAGUCUGCAGGAGCGAAUGAAGUUCGCAUUUGAGG .((((((((((.((((...(((((((((((((((.........(((((....)))))..)))))))))))))))...)))).)))))...))).)).((((((...))))))........ ( -50.00) >DroEre_CAF1 16152 120 + 1 ACCGCAUUUCGAUGAAUAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGCGUGUUCUUCACCGGAAAUCUGCAGGAGCGAAUAAAGUUCGCAUUCGAGG .((((((((((.((((...(((((((((((((((.........(((((....)))))..)))))))))))))))...)))).)))))...))).(((((((((...)))))).)))..)) ( -52.60) >consensus ACCGCAUUUCGAUGAACAUCACGCAGGACGGCCGAUCAGUCUCUCCAGAAGCCUGGAUGCGGCUGUUCUGUGUGUUCUUCAACGGAAGUCUGCAGGAGCGAAUGAAGUUCGCAUUUGAGG .((((((((((.((((...(((((((((((((((.........(((((....)))))..)))))))))))))))...)))).)))))...))).)).((((((...))))))........ (-48.60 = -48.48 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:12 2006