
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,005,888 – 18,006,020 |
| Length | 132 |
| Max. P | 0.998143 |

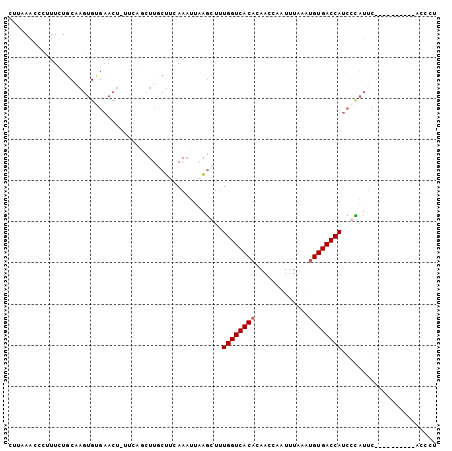
| Location | 18,005,888 – 18,005,982 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 65.30 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -9.55 |
| Energy contribution | -9.79 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18005888 94 + 27905053 GUUAAGCCCUUUCUGCAAGUGUGAACUGUUCCAUUUUCUUAAAAUUCAGCUUUGGUCACACAACCAGUUUUAGUGUGACCAUCCCAUUC----------ACCCU .....((.......)).((.((((((((..................)))...(((((((((...........))))))))).....)))----------)).)) ( -18.17) >DroSim_CAF1 50559 81 + 1 CUUAAACCCUUUCUU-------------UUAAGGUUUCUACAAAUUCGACUUUGGUCACACACCCAGUUUAAAUGUGACCAUCCUGUUA----------ACCCU .......((((....-------------..)))).............(((..((((((((.............))))))))....))).----------..... ( -14.02) >DroEre_CAF1 50120 98 + 1 AUUAAACCCCUUUUGCAAGUGUGAACA------CUUGCUUCAACUUAAGCUCUGGUCACACAACAAAUUUAAAUGUGACCAUCGCAUUCGGAUAAAAGUACCCU ........((....(((((((....))------)))))..........((..((((((((.............))))))))..))....))............. ( -26.62) >DroYak_CAF1 51286 98 + 1 CUUAAAUCCUUUCUGCUAGUGCGAACUCUUCAGCUUGCUUCAAAUUAAGCUUUGGUCACUCAACCCAUUUAAAUGUGACCAUCGUAUGCGGAUAAAAG------ ...........(((((..((((((.......((((((........)))))).(((((((...............))))))))))))))))))......------ ( -25.66) >consensus CUUAAACCCUUUCUGCAAGUGUGAACU_UUCAGCUUGCUUCAAAUUAAGCUUUGGUCACACAACCAAUUUAAAUGUGACCAUCCCAUUC__________ACCCU ....................................................((((((((.............))))))))....................... ( -9.55 = -9.79 + 0.25)


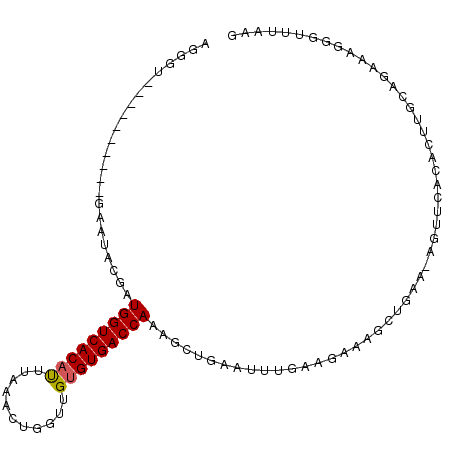
| Location | 18,005,888 – 18,005,982 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.30 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18005888 94 - 27905053 AGGGU----------GAAUGGGAUGGUCACACUAAAACUGGUUGUGUGACCAAAGCUGAAUUUUAAGAAAAUGGAACAGUUCACACUUGCAGAAAGGGCUUAAC .((((----------(.......(((((((((...........))))))))).(((((..(((((......))))))))))..)))))((.......))..... ( -21.60) >DroSim_CAF1 50559 81 - 1 AGGGU----------UAACAGGAUGGUCACAUUUAAACUGGGUGUGUGACCAAAGUCGAAUUUGUAGAAACCUUAA-------------AAGAAAGGGUUUAAG .....----------........(((((((((...........)))))))))...............(((((((..-------------.....)))))))... ( -17.80) >DroEre_CAF1 50120 98 - 1 AGGGUACUUUUAUCCGAAUGCGAUGGUCACAUUUAAAUUUGUUGUGUGACCAGAGCUUAAGUUGAAGCAAG------UGUUCACACUUGCAAAAGGGGUUUAAU .(((((....)))))((((((..(((((((((...........)))))))))..))..........(((((------((....))))))).......))))... ( -31.90) >DroYak_CAF1 51286 98 - 1 ------CUUUUAUCCGCAUACGAUGGUCACAUUUAAAUGGGUUGAGUGACCAAAGCUUAAUUUGAAGCAAGCUGAAGAGUUCGCACUAGCAGAAAGGAUUUAAG ------(((..((((((......(((((((..((((.....)))))))))))..((((......))))..)).......(((((....)).))).))))..))) ( -22.90) >consensus AGGGU__________GAAUACGAUGGUCACAUUUAAACUGGUUGUGUGACCAAAGCUGAAUUUGAAGAAAGCUGAA_AGUUCACACUUGCAGAAAGGGUUUAAG .......................(((((((((...........))))))))).................................................... (-12.16 = -12.22 + 0.06)

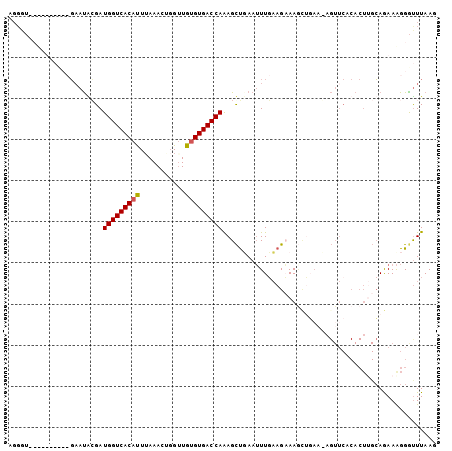
| Location | 18,005,912 – 18,006,020 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 65.22 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -10.47 |
| Energy contribution | -10.35 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18005912 108 + 27905053 ACUGUUCCAUUUUCUUAAAAUUCAGCUUUGGUCACACAACCAGUUUUAGUGUGACCAUCCCAUUC----------ACCCUUUUUUGAGUUUAUCUAAGUUUGAACUCCAAUCGCUCCA .......................(((..(((((((((...........)))))))))........----------..........(((((((........))))))).....)))... ( -22.30) >DroSim_CAF1 50574 99 + 1 ----UUAAGGUUUCUACAAAUUCGACUUUGGUCACACACCCAGUUUAAAUGUGACCAUCCUGUUA----------ACCCUUUCCUA-----UUUUAAGUUCGAACUCCGAUCGUUCCA ----....((.....((......(((..((((((((.............))))))))....))).----------...........-----......(.(((.....))).))).)). ( -14.52) >DroEre_CAF1 50144 112 + 1 ACA------CUUGCUUCAACUUAAGCUCUGGUCACACAACAAAUUUAAAUGUGACCAUCGCAUUCGGAUAAAAGUACCCUUUGUUGAGUAUAUUUAAGUUCGAAUUCCGAUUACUCCA ...------...((((......))))..((((((((.............))))))))(((.((((((((..(((((..(((....)))..)))))..))))))))..)))........ ( -25.02) >DroYak_CAF1 51310 104 + 1 ACUCUUCAGCUUGCUUCAAAUUAAGCUUUGGUCACUCAACCCAUUUAAAUGUGACCAUCGUAUGCGGAUAAAAG--------------UAUAUUAACGUUCGAAUUCCGAUUACUCUA .......((((((........)))))).(((((((...............)))))))..(((..((((.....(--------------(......))........))))..))).... ( -19.38) >consensus ACU_UUCAGCUUGCUUCAAAUUAAGCUUUGGUCACACAACCAAUUUAAAUGUGACCAUCCCAUUC__________ACCCUUU_UUG__UAUAUUUAAGUUCGAACUCCGAUCACUCCA ............................((((((((.............))))))))..........................................(((.....)))........ (-10.47 = -10.35 + -0.13)

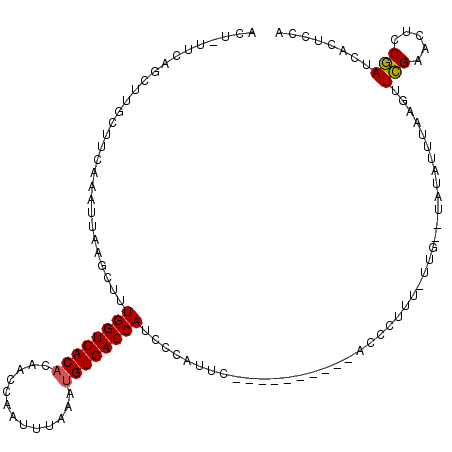
| Location | 18,005,912 – 18,006,020 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 65.22 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -15.00 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18005912 108 - 27905053 UGGAGCGAUUGGAGUUCAAACUUAGAUAAACUCAAAAAAGGGU----------GAAUGGGAUGGUCACACUAAAACUGGUUGUGUGACCAAAGCUGAAUUUUAAGAAAAUGGAACAGU ........((((((((((..((((.....((((......))))----------...)))).(((((((((...........)))))))))....)))))))))).............. ( -26.70) >DroSim_CAF1 50574 99 - 1 UGGAACGAUCGGAGUUCGAACUUAAAA-----UAGGAAAGGGU----------UAACAGGAUGGUCACAUUUAAACUGGGUGUGUGACCAAAGUCGAAUUUGUAGAAACCUUAA---- ........((.((((((((.(((....-----.......(...----------...)....(((((((((...........))))))))))))))))))))...))........---- ( -20.90) >DroEre_CAF1 50144 112 - 1 UGGAGUAAUCGGAAUUCGAACUUAAAUAUACUCAACAAAGGGUACUUUUAUCCGAAUGCGAUGGUCACAUUUAAAUUUGUUGUGUGACCAGAGCUUAAGUUGAAGCAAG------UGU ....(((.(((((.........((((..(((((......)))))..))))))))).)))..(((((((((...........)))))))))..((((......))))...------... ( -29.40) >DroYak_CAF1 51310 104 - 1 UAGAGUAAUCGGAAUUCGAACGUUAAUAUA--------------CUUUUAUCCGCAUACGAUGGUCACAUUUAAAUGGGUUGAGUGACCAAAGCUUAAUUUGAAGCAAGCUGAAGAGU ..((....))...................(--------------(((((....((......(((((((..((((.....)))))))))))..((((......))))..))..)))))) ( -19.20) >consensus UGGAGCAAUCGGAAUUCGAACUUAAAUAUA__CAA_AAAGGGU__________GAAUACGAUGGUCACAUUUAAACUGGUUGUGUGACCAAAGCUGAAUUUGAAGAAAGCUGAA_AGU ..((((.......))))............................................(((((((((...........)))))))))............................ (-15.00 = -14.38 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:30 2006