| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,974,687 – 17,974,794 |
| Length | 107 |
| Max. P | 0.769563 |

| Location | 17,974,687 – 17,974,794 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.79 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -15.00 |
| Energy contribution | -16.87 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
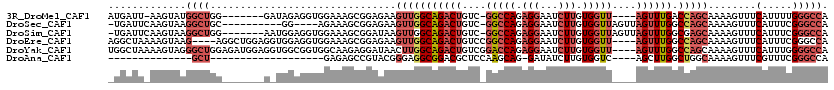
>3R_DroMel_CAF1 17974687 107 - 27905053 AUGAUU-AAGUAUGGCUGG-------GAUAGAGGUGGAAAGCGGAGAAGUUGGCAGACUGUC-GGCCAGAGGAAUCUUGUGGUU----AGUUUGACCAGCAAAAGUUUCAUUUUGGGCCA ......-.....(((((.(-------((..(((.(....).)......((((((((((((.(-.((.(((....))).)).).)----)))))).))))).......))..))).))))) ( -30.10) >DroSec_CAF1 17918 104 - 1 -UGAUUCAAGUAAGGCUGC----------GG----AGAAAGCGGAGAAGUUGGCAGACUGUC-GGCCAGAGGAAUCUUGUGGUUAGUUAGUUUGGCCAGCAAAAGUUUCAUUUCGGGCCA -............((((.(----------((----((.((((......((((((((((((..-(((((.(((...))).)))))...)))))).))))))....))))..))))))))). ( -32.40) >DroSim_CAF1 19368 111 - 1 -UGAUUCAAGUAAGGCUGG-------AAUGGAGGUGGAAAGCGGAUAAGUUGGCAGACUGUC-GGCCAGAGGAAUCUUGUGGUUAGUUAGUUUGGCGAGCAAAAGUUUCAUUUCGGGCCA -............((((((-------(((((((.(....).)......(((.((((((((..-(((((.(((...))).)))))...)))))).)).)))......)))))))).)))). ( -31.80) >DroEre_CAF1 18713 112 - 1 AGGCUAAAAGUAAG----AGGCUGGAGGUGGAGGUGGAAAGCGGAGAAGUUGGCAGACUGUCCGGCCAGAGGAAUCUUGUGGUU----AGUUUGGCCAGCAAAAGUUUCAUUUCGGGCCA ..............----.((((.(((((((((.(.....(((((..((((....)))).)))(((((((..((((....))))----..))))))).))...).))))))))).)))). ( -41.70) >DroYak_CAF1 19027 116 - 1 UGGCUAAAAGUAGGGCUGGAGAUGGAGGUGGCGGUGGCAAGAGGAUAACUUGGCAGACUGUCGGACCAGAGGAAUCUUGUGGUU----AGUUUGGCCAGCAAAAGUUUCAUUUGGGGCCA .((((..((((..((((.....(((...(((((((.((..(((.....))).))..)))))))..(((((..((((....))))----..)))))))).....))))..))))..)))). ( -36.70) >DroAna_CAF1 16954 82 - 1 --------------GCU-------------------GAGAGCCGUACGGGAGGCGGACGCUCCAAGCAG-GAUAUCUUGUGGUC----AGCUUGGCUGGCAAAAGUUUCGUUUCGGGCCA --------------(((-------------------.....((....))((((((((.(((....((((-(....))))).(((----(((...))))))...))))))))))).))).. ( -25.00) >consensus _UGAUU_AAGUAAGGCUGG_______G_UGGAGGUGGAAAGCGGAGAAGUUGGCAGACUGUC_GGCCAGAGGAAUCUUGUGGUU____AGUUUGGCCAGCAAAAGUUUCAUUUCGGGCCA .............((((...............................(((((((((((....(((((.(((...))).)))))....)))))).)))))........(.....))))). (-15.00 = -16.87 + 1.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:19 2006