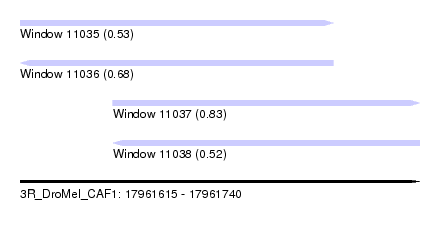
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,961,615 – 17,961,740 |
| Length | 125 |
| Max. P | 0.829745 |

| Location | 17,961,615 – 17,961,713 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -19.83 |
| Energy contribution | -19.25 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17961615 98 + 27905053 CAUGGUUCCUGCUGC----------CACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AGAGGAGCACUCUUCACAGCUCCACA .....((((((((((----------(((....((....)).....))))))))....(((((..........))))).)).)-)).(((((..........)))))... ( -27.60) >DroSec_CAF1 5072 99 + 1 CAUGGUUCCUGCUGC----------CACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCGGAGAGUAGCACUCUUCACAGCUCUACA ....(((...(((((----------(((....((....)).....)))))))).)))...(((.........((.....))((((((.....))))))..)))...... ( -27.90) >DroSim_CAF1 6025 99 + 1 CAUGGUUCCUGCUGC----------CACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCGGAGAGUAGCACUCUUCACAGCUCCACA .........((((((----------..(....)..))))))....((((.(((....(((((..........)))))..(.((((((.....)))))).).))))))). ( -29.30) >DroEre_CAF1 5196 96 + 1 --UGGUUCCUGCUGC----------CACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AGGGGAGUACUCUUCGCAGCUCCACA --.......((((((----------..(....)..))))))....((((.(((....(((((..........)))))..(((-(((((....)))))))).))))))). ( -33.20) >DroYak_CAF1 5462 87 + 1 CAUGGUUGCUGCUGC----------CACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AGAGGAACACUCUUC----------- ..((((.((.(((((----------(((....((....)).....)))))))).......)).)))).....((.....))(-((((.....))))).----------- ( -26.70) >DroAna_CAF1 5040 97 + 1 AAGGGCUGUGGCUGCGCUGAGGCAGCACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AUGCGAGGAGCCUCC----------- ...((((((.(((..((((.(((.........))).))))....))).))))))..(((((((...............))))-))).(((....))).----------- ( -35.76) >consensus CAUGGUUCCUGCUGC__________CACAGAAGCCGCAGCAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG_AGAGGAGCACUCUUCACAGCUCCACA ...((((.(((.((...........))))).))))(((.......(((((((......)))))))......))).......(.((((.....)))).)........... (-19.83 = -19.25 + -0.58)



| Location | 17,961,615 – 17,961,713 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -23.60 |
| Energy contribution | -24.97 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17961615 98 - 27905053 UGUGGAGCUGUGAAGAGUGCUCCUCU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUG----------GCAGCAGGAACCAUG .((((...(((((.(((((.......-))))).)))))....(((((((.......))))))).((((((((((.((....)).----------)))))))))))))). ( -39.10) >DroSec_CAF1 5072 99 - 1 UGUAGAGCUGUGAAGAGUGCUACUCUCCGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUG----------GCAGCAGGAACCAUG (((.((...(((.((((.....)))).)))...)))))....(((((((.......))))))).((((((((((.((....)).----------))))))))))..... ( -36.00) >DroSim_CAF1 6025 99 - 1 UGUGGAGCUGUGAAGAGUGCUACUCUCCGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUG----------GCAGCAGGAACCAUG .((((...(((((.(((((........))))).)))))....(((((((.......))))))).((((((((((.((....)).----------)))))))))))))). ( -39.60) >DroEre_CAF1 5196 96 - 1 UGUGGAGCUGCGAAGAGUACUCCCCU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUG----------GCAGCAGGAACCA-- ..(((...(((((.((((........-.)))).)))))....(((((((.......))))))).((((((((((.((....)).----------)))))))))))))-- ( -36.80) >DroYak_CAF1 5462 87 - 1 -----------GAAGAGUGUUCCUCU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUG----------GCAGCAGCAACCAUG -----------((.(((((.......-))))).))((.....(((((((.......))))))).....((((((.((....)).----------))))))))....... ( -31.60) >DroAna_CAF1 5040 97 - 1 -----------GGAGGCUCCUCGCAU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUGCUGCCUCAGCGCAGCCACAGCCCUU -----------((.(((..((.((((-.(((.((((......))))))).)))).))..))).......((((.((((...((((((...)))))))))).)))))).. ( -36.40) >consensus UGUGGAGCUGUGAAGAGUGCUCCUCU_CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUGCUGCGGCUUCUGUG__________GCAGCAGGAACCAUG ........(((((.(((((........))))).)))))....(((((((.......))))))).((((((((((....................))))))))))..... (-23.60 = -24.97 + 1.36)

| Location | 17,961,644 – 17,961,740 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -16.15 |
| Energy contribution | -15.48 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17961644 96 + 27905053 CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AGAGGAGCACUCUUCACAGCUCCACAGACUCACAGCUGAUGCUAUUUC-------ACCUU ......(((((((......))))))).......((.....))(-((((((((..........)))))..........(((....))).))))-------..... ( -21.60) >DroSec_CAF1 5101 96 + 1 CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCGGAGAGUAGCACUCUUCACAGCUCUACAGACUCACAGCUGAUGCUAUUUC-------ACCU- ....((((((((((....(((((..........)))))..))((((((.....)))))).(((((............))))).)))))))).-------....- ( -23.70) >DroSim_CAF1 6054 96 + 1 CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCGGAGAGUAGCACUCUUCACAGCUCCACAGACUCACAGCUGAUGCUAUUUC-------ACCU- ....((((((((((....(((((..........)))))..))((((((.....)))))).(((((............))))).)))))))).-------....- ( -23.70) >DroEre_CAF1 5223 95 + 1 CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AGGGGAGUACUCUUCGCAGCUCCACAGCUCCACAGCUGAUGCCAUUCC-------ACCU- .....(((((((((....(((((..........)))))..(((-(((((....)))))))).))....(((((....))))).)))))))..-------....- ( -30.90) >DroYak_CAF1 5491 79 + 1 CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AGAGGAACACUCUUC----------------ACAGCUGAUGCUAUUUU-------ACCU- ...(((((((((......(((((..........))))).(((.-.(((((....)))))----------------...)))..)))))))))-------....- ( -19.30) >DroAna_CAF1 5079 86 + 1 CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG-AUGCGAGGAGCCUCC----------------GCAGCUGAUGCCACUUCUGAACGCGGCC- ....((((((((......(((((..........))))).(((.-.((((((....)).)----------------))))))..))))))))............- ( -23.80) >consensus CAAAAAGUGGCAGCCAACAUUGCUACCAAUUAUGCGAUAAGCG_AGAGGAGCACUCUUCACAGCUCCACAGACUCACAGCUGAUGCUAUUUC_______ACCU_ ....((((((((((....(((((..........)))))..))(.((((.....)))).)........................))))))))............. (-16.15 = -15.48 + -0.66)



| Location | 17,961,644 – 17,961,740 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17961644 96 - 27905053 AAGGU-------GAAAUAGCAUCAGCUGUGAGUCUGUGGAGCUGUGAAGAGUGCUCCUCU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG ..(((-------(......)))).((.((((((..(.(((((..........))))).).-.)))))).)).....(((((((.......)))))))....... ( -31.20) >DroSec_CAF1 5101 96 - 1 -AGGU-------GAAAUAGCAUCAGCUGUGAGUCUGUAGAGCUGUGAAGAGUGCUACUCUCCGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG -..((-------((...(((..(((((.((......)).))))).(.((((.....)))).))))..)))).....(((((((.......)))))))....... ( -28.20) >DroSim_CAF1 6054 96 - 1 -AGGU-------GAAAUAGCAUCAGCUGUGAGUCUGUGGAGCUGUGAAGAGUGCUACUCUCCGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG -.(((-------(......))))...(((((....((((((..(((........))).))))))...)))))....(((((((.......)))))))....... ( -28.20) >DroEre_CAF1 5223 95 - 1 -AGGU-------GGAAUGGCAUCAGCUGUGGAGCUGUGGAGCUGCGAAGAGUACUCCCCU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG -..((-------((...(((..(((((....))))).((((.(((.....)))))))...-.)))..)))).....(((((((.......)))))))....... ( -29.70) >DroYak_CAF1 5491 79 - 1 -AGGU-------AAAAUAGCAUCAGCUGU----------------GAAGAGUGUUCCUCU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG -....-------.....(((....)))((----------------((.(((((.......-))))).)))).....(((((((.......)))))))....... ( -21.10) >DroAna_CAF1 5079 86 - 1 -GGCCGCGUUCAGAAGUGGCAUCAGCUGC----------------GGAGGCUCCUCGCAU-CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG -..........((((((((((((((((((----------------(.((....)))))).-.(((.((((......)))))))...))))).)))))))))).. ( -29.00) >consensus _AGGU_______GAAAUAGCAUCAGCUGUGAGUCUGUGGAGCUGUGAAGAGUGCUCCUCU_CGCUUAUCGCAUAAUUGGUAGCAAUGUUGGCUGCCACUUUUUG ...............(((((....))))).............(((((.(((((........))))).)))))....(((((((.......)))))))....... (-18.98 = -19.62 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:14 2006