
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,936,590 – 17,936,683 |
| Length | 93 |
| Max. P | 0.889213 |

| Location | 17,936,590 – 17,936,683 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17936590 93 + 27905053 --GCAG--AGGCC--AAAAAUUAUGCAACUGCAGCCG------CGGCGGUGC-AACAGCUGUUGACACAUGCCGCCAGCGGCCAUUGUUGUAU----GGCCAACACCACU---------- --....--.((((--(.......((((((....((((------(((((((.(-(((....))))......)))))).)))))....)))))))----)))).........---------- ( -38.61) >DroVir_CAF1 9274 104 + 1 --CCAA--AUGCC--AAAAAUUAUGCAACA---CAUG------CGAGUGCGCGAACAGCUGUUGACAUAUUCCACCAGCGGCCAUU-UUGUAGUCCACGCCAACACCAACAGCCGUCGCA --....--.....--........((((...---..))------)).....((((...((((((((((....((......)).....-.))).((........))..)))))))..)))). ( -19.60) >DroPse_CAF1 14408 97 + 1 --GCAA--AGGCC--AAAAAUUAUGCAACUGCAACGGCGAAUGCAACG--GC-AACAGCUGUUGACACAUGCCGCCAGCGGGCAUUGUUGUAU----GGCCAACACCAAU---------- --....--.((((--(..(((.((((..((((..(((((..(((((((--((-....))))))).))..)))))...)))))))).)))...)----)))).........---------- ( -38.10) >DroYak_CAF1 10579 95 + 1 GGGCAG--AGGCC--AAAAAUUAUGCAACUGCAGCCG------CGGCGGUGC-AACAGCUGUUGACACAUGCCGCCAGCGGCCAUUGUUGUAU----GGCCAACACCACU---------- ((((((--..((.--.........))..)))).((.(------(((((((((-(((....)))).))).))))))..))((((((......))----))))....))...---------- ( -40.00) >DroAna_CAF1 13325 95 + 1 --GCAGCAAGGCCCGAAAAAUUGUGCAACUGCAUUGG------UAACA--GC-AACAGCUGUUGACACAUGCCGCCGGCGGGCAUUGUUGUAU----GGCCAACACCACU---------- --(((((((.(((((.....(((.((....((((((.------(((((--((-....))))))).)).)))).)))))))))).))))))).(----((......)))..---------- ( -32.90) >DroPer_CAF1 18299 91 + 1 --GCAG--AGGCC--AAAAAUUAUGCAACUGCAACGG------CAACG--GC-AACAGCUGUUGACACAUGCCGCCAGCGGGCAUUGUUGUAU----GGCCAACACCAAU---------- --....--.((((--(..(((.((((..((((..(((------(((((--((-....))))).......)))))...)))))))).)))...)----)))).........---------- ( -31.01) >consensus __GCAG__AGGCC__AAAAAUUAUGCAACUGCAACGG______CAACG__GC_AACAGCUGUUGACACAUGCCGCCAGCGGCCAUUGUUGUAU____GGCCAACACCAAU__________ .........((((.........(((((((..............(((((..((.....)))))))....((((((....))).))).)))))))....))))................... (-15.00 = -15.50 + 0.50)

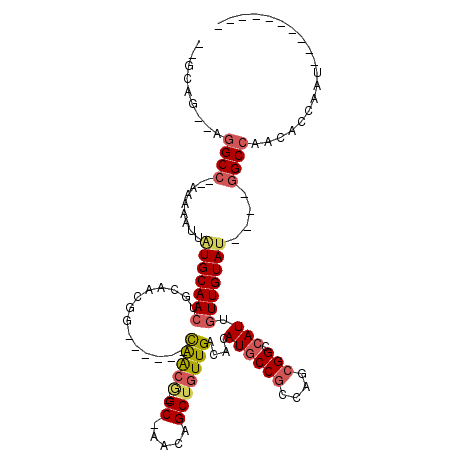
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:57 2006