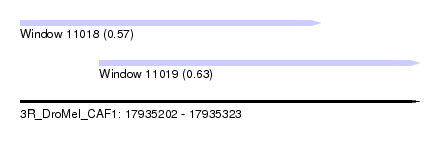
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,935,202 – 17,935,323 |
| Length | 121 |
| Max. P | 0.633905 |

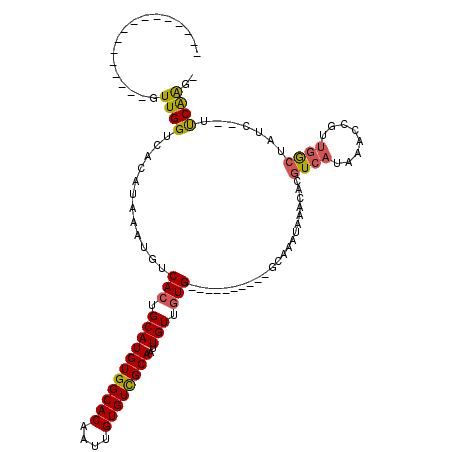
| Location | 17,935,202 – 17,935,293 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -16.02 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17935202 91 + 27905053 ----------------GUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUG----------GCGAAUAAACACGUCAUAAACCGUUGGCUAUC--UUCAAU- ----------------..((((((......((((((.(((((((((((....))))))))..))).)))----------))).......(((........)))))))))..--......- ( -25.50) >DroVir_CAF1 8184 120 + 1 CUGACCAAUGCGAAGCGUUGGUCACAUAAAUGUCACUGCAUGUGGCACGAAUGUGUUGCAAUUGUGUUGAAAUUGGGCCGCAAAUAAACACGUCACAAACGGUUCGCAUUGCUAUCGAGU .(((.(((((((((.((((.((.((.....((((((.....)))))).....(((((....(((((............)))))...))))))).)).)))).)))))))))...)))... ( -36.00) >DroPse_CAF1 12498 92 + 1 ----------------GUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUG----------GCAAAUAAACACGUCAUAAACCGUUGACUAUC--UUCAAGU ----------------..((((((......((((((.(((((((((((....))))))))..))).)))----------))).......(((........)))))))))..--....... ( -26.80) >DroYak_CAF1 9115 91 + 1 ----------------GUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUG----------GCGAAUAAACACGUCAUAAACCGUUGGCCAUC--UUCAAU- ----------------..((((((......((((((.(((((((((((....))))))))..))).)))----------))).......(((........)))))))))..--......- ( -27.80) >DroAna_CAF1 12100 91 + 1 ----------------GUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUG----------GCGAAUAAACACGUCAUAAACCGCCGGGUAUC--UCCAGU- ----------------.((((.........((((((.(((((((((((....))))))))..))).)))----------)))................(((....)))...--.)))).- ( -24.90) >DroPer_CAF1 16416 92 + 1 ----------------GUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUG----------GCAAAUAAACCCGUCAUAAACCGUUGACUAUC--UUCAAGU ----------------..((((((......((((((.(((((((((((....))))))))..))).)))----------)))........((........)).))))))..--....... ( -25.60) >consensus ________________GUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUG__________GCAAAUAAACACGUCAUAAACCGUUGGCUAUC__UUCAAG_ .................((((............(((.(((((((((((....))))))))..))).)))......................((((........)))).......)))).. (-16.02 = -15.92 + -0.11)


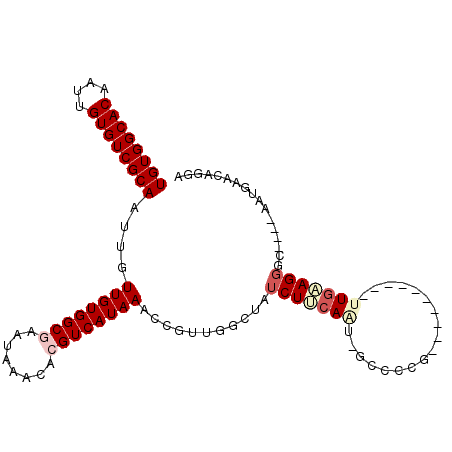
| Location | 17,935,226 – 17,935,323 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -17.97 |
| Energy contribution | -19.05 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17935226 97 + 27905053 UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAAU-GCCUCG----------UUGAAGGGCCUCAAAUGCACUGGA ...(((((....)))))(((.(((((((((((........))))))))......((((..(((((((-(...))----------)))))))))).))).)))...... ( -31.10) >DroPse_CAF1 12522 102 + 1 UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCAAAUAAACACGUCAUAAACCGUUGACUAUCUUCAAGUGCCCCCC--GCCCCGACUGUAGGCU----GCUGACCAAGA ...(((((.....(((((((.....))))))).........((((........)))).........)))))....--(((.........))).----........... ( -22.90) >DroEre_CAF1 14220 97 + 1 UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAAU-GCCUCG----------UUGAAGGGCCUCAAAUGCACAGCA ((((((((....))))))))..((((((((((........))))..........((((..(((((((-(...))----------))))))))))........)))))) ( -31.70) >DroYak_CAF1 9139 97 + 1 UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCCAUCUUCAAU-GACUCG----------UUGAAGGGCCUCAAAUGCACAGGA ((((((((....)))))(((.(((((((((((........))))))))......((((..(((((((-(...))----------)))))))))).))).))))))... ( -33.30) >DroAna_CAF1 12124 96 + 1 UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGCCGGGUAUCUCCAGU-UGCCUG-------UGUUUGGAGUGU----GCUGACCAACA ((((((((....))))))))..((((((((((..................)))))(((((((((((.-......-------...)))))).))----)))...))))) ( -29.87) >DroPer_CAF1 16440 99 + 1 UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCAAAUAAACCCGUCAUAAACCGUUGACUAUCUUCAAGUGCCCCCCCCCCCCCGACUAUAGGCU----GCUGAC----- .(..(((((((((.....)))))))...((((.........((((........))))..........))))...................)).----.)....----- ( -20.41) >consensus UGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAAU_GCCCCG__________UUGAAGGGC____AAUGAACAGGA ((((((((....))))))))....((((((((........))))))))...........(((((((..................)))))))................. (-17.97 = -19.05 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:56 2006