
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,867,331 – 17,867,427 |
| Length | 96 |
| Max. P | 0.942159 |

| Location | 17,867,331 – 17,867,427 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -14.21 |
| Energy contribution | -15.77 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17867331 96 + 27905053 GCCAACGCGCCAAAUUGUGGCCCACCCAGCUACC--AGGUGGCAACGCUGAAUGUUGUUAG---UCA---AUUAAUGCAUUAACUAGCCUCCAUUUGGAAGAAU ((....))((((..(.(((((.......))))).--)..))))....(..((((..(((((---(.(---((......))).))))))...))))..)...... ( -23.90) >DroVir_CAF1 8676 98 + 1 GCCAACGCGCCAAAUA-UGCCUAAACAAGCGAGUGCAAGCGG----GAUGGCUGCUGUUAACAGCGCAUGAUUGACAGGCUUAGA-GCUGCCACCUAACAGAAA ((((.(.(((...((.-(((........))).))....))).----).)))).((((....))))............(((.....-...)))............ ( -24.60) >DroSec_CAF1 8982 96 + 1 GCCAACGCGCCAAAUUGUGGCCCACCCAGCGACC--UGGUGGCAACGCUGAAUAUUGUUAG---UCA---AUUAAUGCAUUAACUAGGCUCCAUUUGGCAGAAU ((....))(((((((.(..(((....(((((.((--....))...)))))......(((((---(((---.....)).))))))..)))..))))))))..... ( -28.50) >DroSim_CAF1 10519 96 + 1 GCCAACGCGCCAAAUUGUGGCCCACCAAGCGACC--AGGUGGCAACGCUAAAUGUUGUUAG---UCA---AUUAAUGCAUUAACUGGGCUUCAUUUGGCAAAAU ((....))(((((((.(.(((((....((((((.--.((((....))))....))))))((---(.(---((......))).)))))))).))))))))..... ( -30.30) >DroEre_CAF1 9607 96 + 1 GCCAACGCGCCAAAUGGUGGCCCACCAAGCGACC--AGGUGGCAACGCUGAAUGUUGUUAG---UCA---AUUAAUUAACUAACUAGGCACUAUUUGGCCGAAU .....((.(((((((((((.((.....((((...--.((((....))))...))))(((((---(.(---(....)).))))))..)))))))))))))))... ( -33.00) >DroYak_CAF1 9699 96 + 1 GCCAACGCGCCAAAUGGUGGCCCACCUAGCGACU--AGGUGGCAACGCUGAAUGUUGUUAG---UCA---AUUAAUGUACUAACUAGACACUAUUUGGCCAAAA ((....))((((((((((((((.((((((...))--))))))).............(((((---(((---.....)).))))))....)))))))))))..... ( -35.10) >consensus GCCAACGCGCCAAAUUGUGGCCCACCAAGCGACC__AGGUGGCAACGCUGAAUGUUGUUAG___UCA___AUUAAUGCACUAACUAGGCUCCAUUUGGCAGAAU (((((...((((.....))))......((((((....((((....))))....))))))...................................)))))..... (-14.21 = -15.77 + 1.56)



| Location | 17,867,331 – 17,867,427 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17867331 96 - 27905053 AUUCUUCCAAAUGGAGGCUAGUUAAUGCAUUAAU---UGA---CUAACAACAUUCAGCGUUGCCACCU--GGUAGCUGGGUGGGCCACAAUUUGGCGCGUUGGC ...(((((....))))).((((((((......))---)))---))).......(((((((..(((((.--.(...)..)))))((((.....))))))))))). ( -34.50) >DroVir_CAF1 8676 98 - 1 UUUCUGUUAGGUGGCAGC-UCUAAGCCUGUCAAUCAUGCGCUGUUAACAGCAGCCAUC----CCGCUUGCACUCGCUUGUUUAGGCA-UAUUUGGCGCGUUGGC .........(((((((((-.....).))))).)))((((((((....)))).((((..----..(((((.((......)).))))).-....)))))))).... ( -28.40) >DroSec_CAF1 8982 96 - 1 AUUCUGCCAAAUGGAGCCUAGUUAAUGCAUUAAU---UGA---CUAACAAUAUUCAGCGUUGCCACCA--GGUCGCUGGGUGGGCCACAAUUUGGCGCGUUGGC .....((((((((..(((((((((((......))---)))---)))....(((((((((..(((....--)))))))))))))))..).)))))))........ ( -35.40) >DroSim_CAF1 10519 96 - 1 AUUUUGCCAAAUGAAGCCCAGUUAAUGCAUUAAU---UGA---CUAACAACAUUUAGCGUUGCCACCU--GGUCGCUUGGUGGGCCACAAUUUGGCGCGUUGGC .....(((((((...(((((((((((......))---)))---))......((..((((..(((....--)))))))..))))))....)))))))........ ( -30.90) >DroEre_CAF1 9607 96 - 1 AUUCGGCCAAAUAGUGCCUAGUUAGUUAAUUAAU---UGA---CUAACAACAUUCAGCGUUGCCACCU--GGUCGCUUGGUGGGCCACCAUUUGGCGCGUUGGC ...(((((((((.(((((..(((((((((....)---)))---)))))..(((..((((..(((....--)))))))..))))).))).))))))).))..... ( -36.40) >DroYak_CAF1 9699 96 - 1 UUUUGGCCAAAUAGUGUCUAGUUAGUACAUUAAU---UGA---CUAACAACAUUCAGCGUUGCCACCU--AGUCGCUAGGUGGGCCACCAUUUGGCGCGUUGGC .....(((((..(((((...((((((.((.....---)))---))))).)))))..((....((((((--((...))))))))((((.....)))))).))))) ( -33.20) >consensus AUUCUGCCAAAUGGAGCCUAGUUAAUGCAUUAAU___UGA___CUAACAACAUUCAGCGUUGCCACCU__GGUCGCUUGGUGGGCCACAAUUUGGCGCGUUGGC .....................................................(((((((..(((((...........)))))((((.....))))))))))). (-14.35 = -14.97 + 0.61)

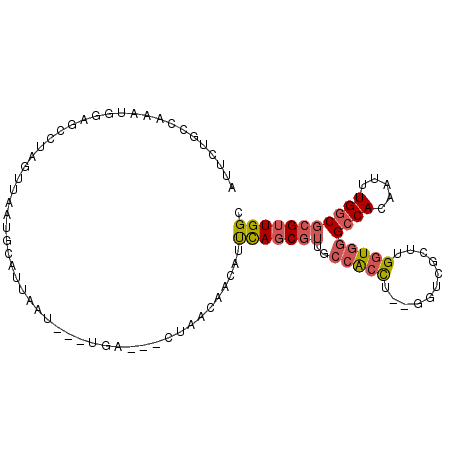

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:37 2006