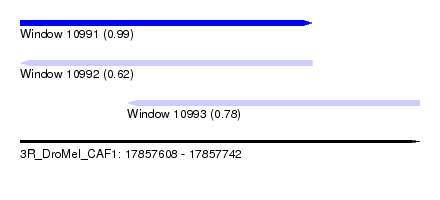
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,857,608 – 17,857,742 |
| Length | 134 |
| Max. P | 0.993626 |

| Location | 17,857,608 – 17,857,706 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17857608 98 + 27905053 GUGGAAAAUAUCGAUUUUGGCCAGCACUACUCGCAUAUGUUUUGACUUGCCAGUGGGGGUGGCUGGGCCA-UUGGGUGGUGGGCCGACCAUCCGUCGAG ..........(((((..((((((((.(((((.(((...((....)).))).))))).....))).)))))-..(((((((......)))))))))))). ( -36.60) >DroSec_CAF1 2154 98 + 1 GUGGAAAAUAUCGAUUUUGGCCAGCACUACUCGCAUAUGUUUUGGCUUGCAAGUGGGGGUGGCUGGGCCA-UAGGGUGGUGGGCGGACCAUCCGUCGAG ..........(((((..((((((((.(.((((.(((.(((........))).))).)))))))).)))))-..(((((((......)))))))))))). ( -38.60) >DroSim_CAF1 2160 98 + 1 GUGGAAAAUAUCGAUUUUGGCCAGCACUACUCGCAUAUGUUUUGGCUUGCAAGUGGGGGUGGAUGGGCCA-UAGGGUGGUGGGCGGACCAUCCGUCGAG ..........(((((..(((((....((((((.(((.(((........))).))).))))))...)))))-..(((((((......)))))))))))). ( -38.50) >DroEre_CAF1 2198 98 + 1 GUGGAAAAUAUCGAUUUUGGCCAGCACUACUCGCAUAUGUUUUGGCUUUACUGCGGGGGUGGCUGGGCCA-UUGGGUGGUGGGCGGACCAUCCGUCGAG ..........(((((..(((((((((((.((((((...((....)).....))))))))).))).)))))-..(((((((......)))))))))))). ( -37.50) >DroYak_CAF1 2206 99 + 1 GUGGAAAAUAUCGAUUUUGGCCAGCACUACUCGCAUAUGUUUUGGCUUGACUGUGGGGGUGGCUGGGCCAAAUGGGUGGUGGGCGGACCAUCCGUCGAU .........((((((((((((((((.(.((((.((((.(((.......))))))).)))))))).))))))).(((((((......))))))))))))) ( -40.00) >consensus GUGGAAAAUAUCGAUUUUGGCCAGCACUACUCGCAUAUGUUUUGGCUUGCCAGUGGGGGUGGCUGGGCCA_UUGGGUGGUGGGCGGACCAUCCGUCGAG ..........(((((..(((((((((((.(((((....((....))......)))))))).))).)))))...(((((((......)))))))))))). (-32.92 = -32.80 + -0.12)



| Location | 17,857,608 – 17,857,706 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -19.96 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17857608 98 - 27905053 CUCGACGGAUGGUCGGCCCACCACCCAA-UGGCCCAGCCACCCCCACUGGCAAGUCAAAACAUAUGCGAGUAGUGCUGGCCAAAAUCGAUAUUUUCCAC .((((.((.((((......)))).))..-(((((.(((.((....(((.(((.((......)).))).))).))))))))))...)))).......... ( -27.80) >DroSec_CAF1 2154 98 - 1 CUCGACGGAUGGUCCGCCCACCACCCUA-UGGCCCAGCCACCCCCACUUGCAAGCCAAAACAUAUGCGAGUAGUGCUGGCCAAAAUCGAUAUUUUCCAC .((((.((.((((......)))).))..-(((((.(((.((....(((((((............))))))).))))))))))...)))).......... ( -28.20) >DroSim_CAF1 2160 98 - 1 CUCGACGGAUGGUCCGCCCACCACCCUA-UGGCCCAUCCACCCCCACUUGCAAGCCAAAACAUAUGCGAGUAGUGCUGGCCAAAAUCGAUAUUUUCCAC .((((.((.((((......)))).))..-(((((....(((....(((((((............))))))).)))..)))))...)))).......... ( -25.20) >DroEre_CAF1 2198 98 - 1 CUCGACGGAUGGUCCGCCCACCACCCAA-UGGCCCAGCCACCCCCGCAGUAAAGCCAAAACAUAUGCGAGUAGUGCUGGCCAAAAUCGAUAUUUUCCAC .((((.((.((((......)))).))..-(((((.(((.((..(((((................)))).)..))))))))))...)))).......... ( -28.09) >DroYak_CAF1 2206 99 - 1 AUCGACGGAUGGUCCGCCCACCACCCAUUUGGCCCAGCCACCCCCACAGUCAAGCCAAAACAUAUGCGAGUAGUGCUGGCCAAAAUCGAUAUUUUCCAC (((((.((.((((......)))).)).(((((((.(((.((..((.((................)).).)..)))))))))))).)))))......... ( -25.59) >consensus CUCGACGGAUGGUCCGCCCACCACCCAA_UGGCCCAGCCACCCCCACUGGCAAGCCAAAACAUAUGCGAGUAGUGCUGGCCAAAAUCGAUAUUUUCCAC .((((.((.((((......)))).))...(((((.(((.((....(((.(((............))).))).))))))))))...)))).......... (-19.96 = -21.00 + 1.04)



| Location | 17,857,644 – 17,857,742 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17857644 98 - 27905053 AGUACGCAUGCGCAGUGCAUGUGAAAUGGCAAACAACUCGACGGAUGGUCGGCCCACCACCCAA-UGGCCCAGCCACCCCCACUGGCAAGUCAAAACAU ....(((((((.....)))))))...((((.........(((.....)))((((..........-.))))..((((.......))))..))))...... ( -26.20) >DroSec_CAF1 2190 98 - 1 AGUCCGCAUGCGCAGUGCAUGUGAAAUGGCAAACAACUCGACGGAUGGUCCGCCCACCACCCUA-UGGCCCAGCCACCCCCACUUGCAAGCCAAAACAU ....(((((((.....)))))))...((((............((.((((......)))).))..-((((...)))).............))))...... ( -25.60) >DroSim_CAF1 2196 98 - 1 AGUCCGCAUGCGCAGUGCAUGUGAAAUGGCAAACAACUCGACGGAUGGUCCGCCCACCACCCUA-UGGCCCAUCCACCCCCACUUGCAAGCCAAAACAU ....(((((((.....)))))))...((((...(.....)..((((((.(((............-))).))))))..............))))...... ( -25.60) >DroEre_CAF1 2234 98 - 1 AGUCCGCAUGCGCAGUGCAUGUGAAAUGGCAAACAACUCGACGGAUGGUCCGCCCACCACCCAA-UGGCCCAGCCACCCCCGCAGUAAAGCCAAAACAU ....(((((((.....)))))))...((((.....(((((..((.((((......)))).))..-((((...))))....)).)))...))))...... ( -26.90) >DroYak_CAF1 2242 99 - 1 AGUCCGCAUGCGCAGUGCAUGUGAAGUGGCAAACAAAUCGACGGAUGGUCCGCCCACCACCCAUUUGGCCCAGCCACCCCCACAGUCAAGCCAAAACAU ....(((((((.....)))))))...((((............((.((((......)))).))...((((...)))).............))))...... ( -25.90) >consensus AGUCCGCAUGCGCAGUGCAUGUGAAAUGGCAAACAACUCGACGGAUGGUCCGCCCACCACCCAA_UGGCCCAGCCACCCCCACUGGCAAGCCAAAACAU ....(((((((.....)))))))...((((............((.((((......)))).))...((((...)))).............))))...... (-24.30 = -24.34 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:31 2006