
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,852,625 – 17,852,736 |
| Length | 111 |
| Max. P | 0.845178 |

| Location | 17,852,625 – 17,852,736 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -44.33 |
| Consensus MFE | -24.97 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17852625 111 + 27905053 UUUGCAUC---UCCAUGCUUCUGAUGCAUGGGGCAUUCUGGUGGCGGGGCACCACUGCCGCUCAUGUACUUGGCGUGGUCCACCGGCACACUCUUCGUGGCCUCCAUUUGCACU ..((((.(---(((((((.......)))))))).....(((.(((((((.(((((.((((..........))))))))))).))....(((.....)))))).)))..)))).. ( -44.20) >DroPse_CAF1 769 114 + 1 UUGUUAUCGCCCCCCUGCUUCUGGUGCAUGGGGCACUCCGGCGGCGGUGAGCCACCUGCGGCCAUGUGCUUGGCAUGAUCAGCCGGCACACCUUUCGGCGCUUCCAUCUGUACG .((..(.(((((((((((.......))).))))......(((.((((((...)))).)).))).((((((.(((.......)))))))))......)))).)..))........ ( -43.10) >DroEre_CAF1 770 111 + 1 UUUGAAUC---UCCAUGCUUCUGAUGCAUGGGGCAUUCUGGUGGCGGGGAACCACCAGCGCCCAUGUACUUGGCGUGGUCCACCGGCACACUCUUAGUGGCCUCCAUUUGCACA ...(((((---(((((((.......)))))))).)))).(((((.((....))((((.((((.........)))))))))))))(((.(((.....))))))............ ( -45.50) >DroWil_CAF1 705 108 + 1 UGUUCGUC------UUGUUUCUUAUGCAUGGGACAUUCGGGUGGUGGUGAUCCACCGCCAGCCAUGUGCUUGGCAUGAUCAGCUGGCGUACUCUUGGUGGCCUCCAUCUGUACG .....(((------..(((..((((((..((.((((..((.((((((((...)))))))).)))))).))..))))))..))).)))((((....(((((...))))).)))). ( -41.40) >DroYak_CAF1 793 111 + 1 CUCGCAUC---UCCAUGCUUCUGGUGCAUGGGGCAUUCUGGUGGCGGGGAACCACCACCGCUCAUGUACUUGGCGUGGUCUACCGGCACACUCUUCGUGGCCUCCAUUUGCACU ...(((..---.(((((((...((((((((((......((((((.((....)).)))))))))))))))).)))))))......(((.(((.....))))))......)))... ( -48.80) >DroPer_CAF1 1720 114 + 1 UUGUUAUCGCCCCCCUGCUUCUGGUGCAUGGGGCACUCCGGCGGCGGUGAGCCACCUGCGGCCAUGUGCUUGGCAUGAUCGGCCGGCACACCUUUCGGCGCUUCCAUCUGUACG .((..(.(((((((((((.......))).))))......(((.((((((...)))).)).))).((((((.(((.......)))))))))......)))).)..))........ ( -43.00) >consensus UUUGCAUC___UCCAUGCUUCUGAUGCAUGGGGCAUUCUGGUGGCGGGGAACCACCACCGCCCAUGUACUUGGCAUGAUCAACCGGCACACUCUUCGUGGCCUCCAUCUGCACG ........................((((((((((...(((((((.((....)).))......(((((.....)))))....)))))..(((.....)))))).)))..)))).. (-24.97 = -24.25 + -0.72)



| Location | 17,852,625 – 17,852,736 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -43.37 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17852625 111 - 27905053 AGUGCAAAUGGAGGCCACGAAGAGUGUGCCGGUGGACCACGCCAAGUACAUGAGCGGCAGUGGUGCCCCGCCACCAGAAUGCCCCAUGCAUCAGAAGCAUGGA---GAUGCAAA ..((((..(((.((((((.....))).)))(((((((((((((............))).)))))...))))).))).......((((((.......)))))).---..)))).. ( -43.80) >DroPse_CAF1 769 114 - 1 CGUACAGAUGGAAGCGCCGAAAGGUGUGCCGGCUGAUCAUGCCAAGCACAUGGCCGCAGGUGGCUCACCGCCGCCGGAGUGCCCCAUGCACCAGAAGCAGGGGGGCGAUAACAA .((.(((.(((..(((((....))))).))).)))....((((..((((.((((.((.((((...)))))).))))..))))(((.(((.......))).)))))))...)).. ( -48.80) >DroEre_CAF1 770 111 - 1 UGUGCAAAUGGAGGCCACUAAGAGUGUGCCGGUGGACCACGCCAAGUACAUGGGCGCUGGUGGUUCCCCGCCACCAGAAUGCCCCAUGCAUCAGAAGCAUGGA---GAUUCAAA (((((.......((((((.....))).)))((((.....))))..))))).((((.((((((((.....))))))))...))))(((((.......)))))..---........ ( -46.90) >DroWil_CAF1 705 108 - 1 CGUACAGAUGGAGGCCACCAAGAGUACGCCAGCUGAUCAUGCCAAGCACAUGGCUGGCGGUGGAUCACCACCACCCGAAUGUCCCAUGCAUAAGAAACAA------GACGAACA .......((((.(((............((((((((....(((...)))..))))))))(((((....)))))........))))))).............------........ ( -28.70) >DroYak_CAF1 793 111 - 1 AGUGCAAAUGGAGGCCACGAAGAGUGUGCCGGUAGACCACGCCAAGUACAUGAGCGGUGGUGGUUCCCCGCCACCAGAAUGCCCCAUGCACCAGAAGCAUGGA---GAUGCGAG .((((...(((.((((((.....))).)))((....))...))).))))....((((((((((....))))))))........((((((.......)))))).---...))... ( -43.80) >DroPer_CAF1 1720 114 - 1 CGUACAGAUGGAAGCGCCGAAAGGUGUGCCGGCCGAUCAUGCCAAGCACAUGGCCGCAGGUGGCUCACCGCCGCCGGAGUGCCCCAUGCACCAGAAGCAGGGGGGCGAUAACAA .............(((((....))))).(((((((..((((.......))))..))..(((((....))))))))))..((((((.(((.......)))..))))))....... ( -48.20) >consensus CGUACAAAUGGAGGCCACGAAGAGUGUGCCGGCGGACCACGCCAAGCACAUGGCCGCAGGUGGUUCACCGCCACCAGAAUGCCCCAUGCACCAGAAGCAUGGA___GAUGAAAA .......((((.(((........((((((.(((.......)))..)))))).......((((((.....)))))).....)))))))........................... (-23.84 = -23.98 + 0.14)

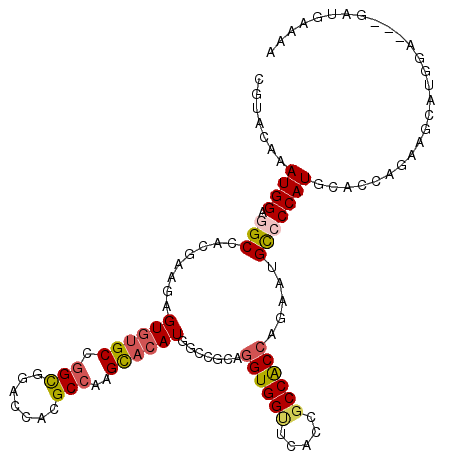

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:27 2006