
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,851,237 – 17,851,327 |
| Length | 90 |
| Max. P | 0.991995 |

| Location | 17,851,237 – 17,851,327 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
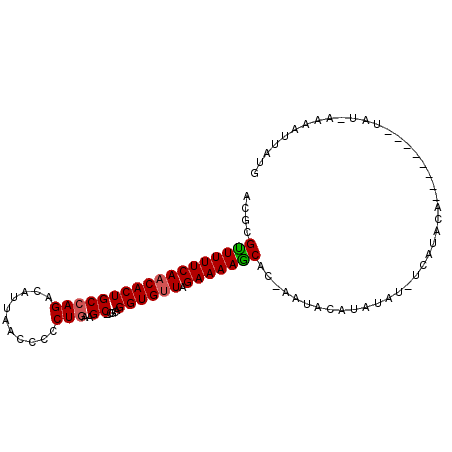
>3R_DroMel_CAF1 17851237 90 + 27905053 ACGCGCUUUUCAACACUGCCAGACAUUAACCCCCUGAAGC-CGCAGGUGUUAGAAAAGCAU-CAUACAUAUAA-ACAUACA--------UAU-AAUGUUAUG ....((((((((((((((((((...........)))....-.))).))))).)))))))..-..........(-((((...--------...-.)))))... ( -18.60) >DroSim_CAF1 6002 93 + 1 ACGCGUUUUUCAACACUGCCAGACAUUAACCCCCUAAAGC-CACAGGUGUUAGAAAAACGCAAAUACAUAUAUUUCAUAAA--------UAUGAAAAAUAUA ..(((((((((((((((....(.(.(((......))).))-....)))))).)))))))))......((((.((((((...--------.)))))).)))). ( -22.30) >DroEre_CAF1 5703 75 + 1 ACGCGUUUUUCAACACUGCCAGACAUUAACCCCCUGAAGC-CGCAGGUGCUAGAAAAGCACGAAUA-------------------------U-GAUAUUUCG .((.(((((((.((.(((((((...........)))....-.))))))....))))))).))....-------------------------.-......... ( -14.10) >DroYak_CAF1 5717 101 + 1 ACGCGUUUUUCAACACUGCCAGACAUUAACCCCCUGAAGCCCGCAGGUGUUGGAAAAGCACCAAUACAUACAUUUCAUACAAUUGUAGGUAU-AAAGUUUUG ..((.(((((((((((((((((...........)))......))).)))))))))))))....((((.((((...........)))).))))-......... ( -21.30) >consensus ACGCGUUUUUCAACACUGCCAGACAUUAACCCCCUGAAGC_CGCAGGUGUUAGAAAAGCAC_AAUACAUAUAU_UCAUACA________UAU_AAAAUUAUG ....((((((((((((((((((...........)))..)).....)))))).)))))))........................................... (-12.25 = -12.38 + 0.12)


| Location | 17,851,237 – 17,851,327 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -18.10 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17851237 90 - 27905053 CAUAACAUU-AUA--------UGUAUGU-UUAUAUGUAUG-AUGCUUUUCUAACACCUGCG-GCUUCAGGGGGUUAAUGUCUGGCAGUGUUGAAAAGCGCGU ......(((-(((--------(((((..-..)))))))))-))(((((((.(((((.(((.-....((((.(.....).))))))))))))))))))).... ( -25.00) >DroSim_CAF1 6002 93 - 1 UAUAUUUUUCAUA--------UUUAUGAAAUAUAUGUAUUUGCGUUUUUCUAACACCUGUG-GCUUUAGGGGGUUAAUGUCUGGCAGUGUUGAAAAACGCGU (((((.((((((.--------...)))))).))))).....(((((((((.(((((.((((-((.((((....)))).)))..))))))))))))))))).. ( -26.80) >DroEre_CAF1 5703 75 - 1 CGAAAUAUC-A-------------------------UAUUCGUGCUUUUCUAGCACCUGCG-GCUUCAGGGGGUUAAUGUCUGGCAGUGUUGAAAAACGCGU .........-.-------------------------....((((.(((((.(((((.(((.-....((((.(.....).))))))))))))))))).)))). ( -17.90) >DroYak_CAF1 5717 101 - 1 CAAAACUUU-AUACCUACAAUUGUAUGAAAUGUAUGUAUUGGUGCUUUUCCAACACCUGCGGGCUUCAGGGGGUUAAUGUCUGGCAGUGUUGAAAAACGCGU ....(((..-((((.((((.((.....)).)))).)))).)))((((((.((((((.(((((((..............)))).))))))))).)))).)).. ( -22.64) >consensus CAAAACAUU_AUA________UGUAUGA_AUAUAUGUAUU_GUGCUUUUCUAACACCUGCG_GCUUCAGGGGGUUAAUGUCUGGCAGUGUUGAAAAACGCGU .........................................(((((((((.(((((.(((......((((.(.....).))))))))))))))))))))).. (-18.10 = -17.98 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:25 2006