
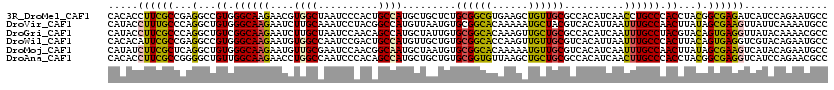
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,842,811 – 17,842,931 |
| Length | 120 |
| Max. P | 0.550769 |

| Location | 17,842,811 – 17,842,931 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
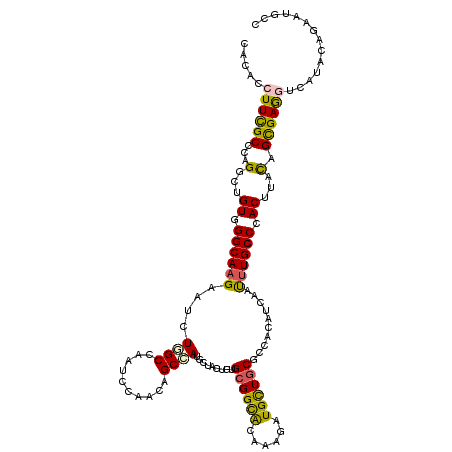
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -22.55 |
| Energy contribution | -21.75 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17842811 120 - 27905053 CACACCUUCGCCGAGGCCGUGGGCAAGAACGUGGCUAAUCCCACUGCCAUGCUGCUCUGCGGCGUGAAGCUGUUGCGCCACAUCAACCUGCCCACCUACGGCGAGAUCAUCCAGAAUGCC ......(((((((.((..(((((((.(...(((((............((((((((...))))))))..((....))))))).....).))))))))).)))))))............... ( -45.60) >DroVir_CAF1 139070 120 - 1 CAUACCUUUGCCCAGGCUGUGGGCAAGAAUCUUGCAAAUCCUACGGCCAUGUUAAUGUGCGGCACAAAAAUGCUACGUCACAUUAAUUUGCCAACUUAUAGCGAAGUUAUUCAAAAUGCC .(((.((((((...(((((((((((((...))))).....))))))))..((((((((((((((......)))..)).))))))))).............)))))).))).......... ( -34.10) >DroGri_CAF1 129627 120 - 1 CAUACCUUCGCCCAGGCUGUCGGCAAGAAUCUUGCUAAUCCAACAGCCAUGCUAUUGUGCGGCACAAAGUUGCUGCGCCACAUCAAUUUGCCUACGUACAGUGAGGUUAUACAAAACGCC .(((.((((((...((((((.((((((...))))))......)))))).......((((((.....((((((.((.....)).)))))).....)))))))))))).))).......... ( -33.10) >DroWil_CAF1 168630 120 - 1 CACACAUUCGCCGAGGCCGUGGGCAAGAAUGUGGCCAAUCCGACUGCCAUGUUGCUGUGCGGCACCAAGUUGUUGCGUCACAUUAAUUUGCCCACUUACAGUGAGGUCGUACAGAAUGCC ....(((((.....((((((((((((.(((((((((((..(((((....((((((...))))))...)))))))).))))))))...))))))))((.....)))))).....))))).. ( -43.90) >DroMoj_CAF1 158773 120 - 1 CAUAUCUUCGCUCAGGCUGUGGGCAAGAAUGUUGCGAAUCCAACGGCAAUGCUAAUGUGCGGCACAAAAAUGUUGCGUCACAUCAAUUUGCCAACUUAUAGCGAAGUCAUACAGAAUGCC .....(((((((......((.((((((...((((......))))(((...((....))((((((......))))))))).......)))))).))....))))))).............. ( -29.10) >DroAna_CAF1 138934 120 - 1 CACACCUUCGCCGGGGCUGUUGGCAAGAACCUGGCCAAUCCCACAGCCAUGCUGCUGUGCGGUGUUAAGCUGCUGCGCCACAUCAACUUGCCCACCUACGGCGAGGUCAUCCAGAACGCC ...((((.((((((((.((..((((((....((((....((((((((......)))))).))......((....))))))......))))))))))).)))))))))............. ( -43.80) >consensus CACACCUUCGCCCAGGCUGUGGGCAAGAAUCUGGCCAAUCCAACAGCCAUGCUACUGUGCGGCACAAAGAUGCUGCGCCACAUCAAUUUGCCCACUUACAGCGAGGUCAUACAGAAUGCC .....((((((...(...((.((((((....((((..........)))).........((((((......))))))..........)))))).))...).)))))).............. (-22.55 = -21.75 + -0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:18 2006