
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,794,031 – 17,794,121 |
| Length | 90 |
| Max. P | 0.988312 |

| Location | 17,794,031 – 17,794,121 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.84 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
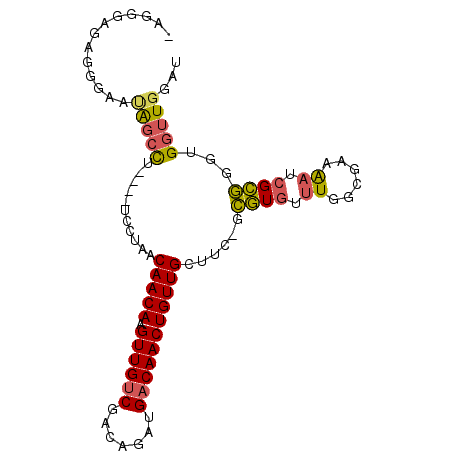
>3R_DroMel_CAF1 17794031 90 + 27905053 AUCCAACCACCCGCGAUUUUCGCCAAACACGC-GAAGCAACAGUUGUCAUCUGUCGACAACUUGUUGUUAGGA----AAGCUAUUCCCUCUCCCU- .(((.......((((..............)))-).(((((((((((((.......)))))).))))))).)))----..................- ( -22.04) >DroSim_CAF1 82189 80 + 1 AUCCAACCACCCGCGAUUUUCGCAAAACACGC-GAAGCAACAGUUGUCAUCUGUCGACAACUUGUUGUUAGGA----AAGCUAUU----------- .(((........(((.....))).........-..(((((((((((((.......)))))).))))))).)))----........----------- ( -22.50) >DroEre_CAF1 88250 90 + 1 AUCCAACCACCCGCGAUUUUCGCCAAACACGC-GAAGCAACAGUUGUCAUCUGUCGACAACUUGUUGUUAGGA----AGGCCAUUCCCUCUCCCU- ...........((((..............)))-).(((((((((((((.......)))))).))))))).(((----(((......))).)))..- ( -22.94) >DroYak_CAF1 88844 94 + 1 AUCCAACCACCCGCGAUUUUUGCCAAACACGC-GAAGCAACAGUUGUCAUCUGUCGACAACUUGUUGUUAGGAAGAAAGGCUUUCCCCUCUCCCA- .(((.......((((..............)))-).(((((((((((((.......)))))).))))))).))).((.(((......))).))...- ( -24.44) >DroAna_CAF1 90055 89 + 1 ACCCACUCACCAACC--CUCCACCACCCAUCCAGCAGCAACAGUUGGCCGC-GGCGACAACUUGUUGUUCCAA----AGCUCGUGCUCUCACUCUC ...............--................(.(((((((((((((...-.))..)))).))))))).)..----(((....)))......... ( -14.40) >consensus AUCCAACCACCCGCGAUUUUCGCCAAACACGC_GAAGCAACAGUUGUCAUCUGUCGACAACUUGUUGUUAGGA____AGGCUAUUCCCUCUCCCU_ ......((....(((.....)))............(((((((((((((.......)))))).))))))).))........................ (-14.82 = -15.38 + 0.56)



| Location | 17,794,031 – 17,794,121 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.84 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.72 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17794031 90 - 27905053 -AGGGAGAGGGAAUAGCUU----UCCUAACAACAAGUUGUCGACAGAUGACAACUGUUGCUUC-GCGUGUUUGGCGAAAAUCGCGGGUGGUUGGAU -.(((((((.......)))----))))..((((.((((((((.....))))))))...(((.(-(((.((((.....))))))))))).))))... ( -27.50) >DroSim_CAF1 82189 80 - 1 -----------AAUAGCUU----UCCUAACAACAAGUUGUCGACAGAUGACAACUGUUGCUUC-GCGUGUUUUGCGAAAAUCGCGGGUGGUUGGAU -----------..(((((.----(((...(((((.(((((((.....)))))))))))).(((-(((.....))))))......))).)))))... ( -25.10) >DroEre_CAF1 88250 90 - 1 -AGGGAGAGGGAAUGGCCU----UCCUAACAACAAGUUGUCGACAGAUGACAACUGUUGCUUC-GCGUGUUUGGCGAAAAUCGCGGGUGGUUGGAU -.....(((((.....)))----))(((((((((.(((((((.....)))))))))))(((.(-(((.((((.....))))))))))).))))).. ( -26.80) >DroYak_CAF1 88844 94 - 1 -UGGGAGAGGGGAAAGCCUUUCUUCCUAACAACAAGUUGUCGACAGAUGACAACUGUUGCUUC-GCGUGUUUGGCAAAAAUCGCGGGUGGUUGGAU -.((((((((((....))))))))))...((((.((((((((.....))))))))...(((.(-(((.((((.....))))))))))).))))... ( -33.70) >DroAna_CAF1 90055 89 - 1 GAGAGUGAGAGCACGAGCU----UUGGAACAACAAGUUGUCGCC-GCGGCCAACUGUUGCUGCUGGAUGGGUGGUGGAG--GGUUGGUGAGUGGGU .......(((((....)))----)).((.(((....))))).((-((.(((((((.(..(..((.....))..)..)..--)))))))..)))).. ( -28.30) >consensus _AGGGAGAGGGAAUAGCCU____UCCUAACAACAAGUUGUCGACAGAUGACAACUGUUGCUUC_GCGUGUUUGGCGAAAAUCGCGGGUGGUUGGAU .............(((((...........(((((.((((((.......)))))))))))......((((.((......)).))))...)))))... (-14.92 = -14.72 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:09 2006