
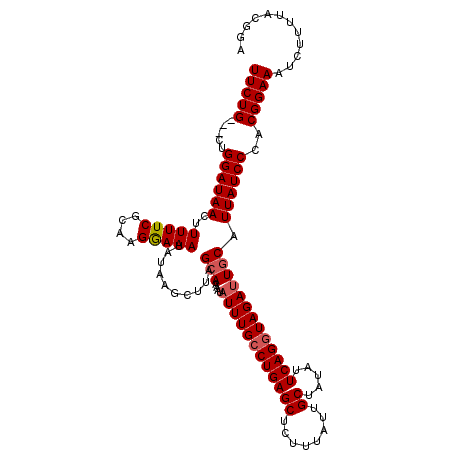
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,789,075 – 17,789,226 |
| Length | 151 |
| Max. P | 0.999174 |

| Location | 17,789,075 – 17,789,190 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
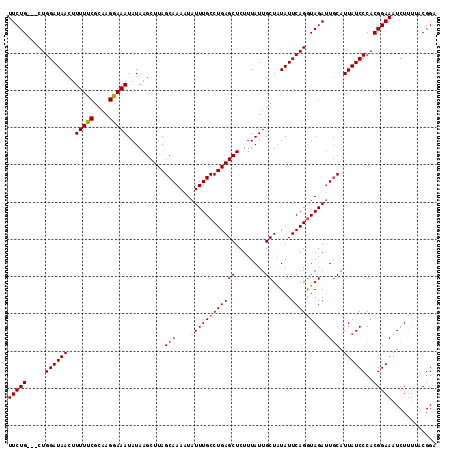
>3R_DroMel_CAF1 17789075 115 - 27905053 UUCUGCUGCUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAUUCAGGUAGAUUGCAUUAUCCCACGGAAAUCUUUUACGGA (((((.((..((((((..(((((....)))))..........(((....((((((((((((........)).....))))))))))))).)))))))))))))............ ( -28.50) >DroSec_CAF1 77499 112 - 1 UUCUG---CUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAUUCAGGUAGAUUGCAUUAUCCCACGGAAAUCUUUUACGGA (((((---..((((((..(((((....)))))..........(((....((((((((((((........)).....))))))))))))).))))))..)))))............ ( -29.60) >DroSim_CAF1 78239 112 - 1 UUCUG---CUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAUUCAGGUAGAUUGCAUUAUCCCACGGAAAUCUUUUACGGA (((((---..((((((..(((((....)))))..........(((....((((((((((((........)).....))))))))))))).))))))..)))))............ ( -29.60) >DroEre_CAF1 83252 112 - 1 UUCUG---CUGGAUAACUUUUUCGCAAGAAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAUUCAGGUAGAGUCCAUUAUCCCACGGAAAUCUCUAGCGGC ..(((---(((((.....(((((....)))))..................(((((((((((........)).....))))))))).(((..........)))....)))))))). ( -27.60) >DroYak_CAF1 83781 112 - 1 UUCUG---CUGGAUAACUUUUUCGCAAGAAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUGUAUUCAGAUAGAUUGCAUUAUCCCACGGAAAUCUUGCACGGC .((((---(((((((...(((((....)))))....((((((((((.....))).)))))))...........))))))).)))).((((...(((...))).....)))).... ( -27.80) >consensus UUCUG___CUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAUUCAGGUAGAUUGCAUUAUCCCACGGAAAUCUUUUACGGA (((((.....((((((..(((((....)))))..........(((....((((((((((((........)).....))))))))))))).))))))..)))))............ (-24.46 = -24.82 + 0.36)

| Location | 17,789,114 – 17,789,226 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -22.47 |
| Energy contribution | -22.23 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
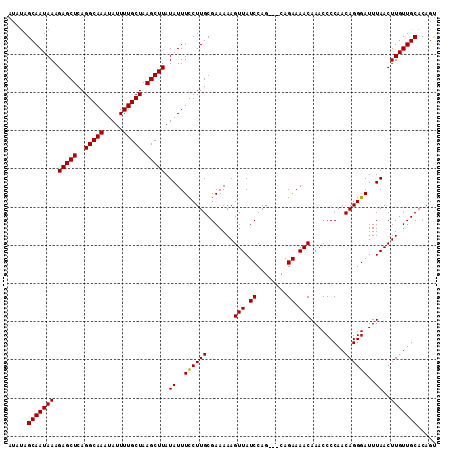
>3R_DroMel_CAF1 17789114 112 + 27905053 AUAUAGCAAUAAAGAGCUCAGGCAAAUAUUUUGCUAAGCUUAUAUUUCCUUGCGAAAAAGUUAUCCAGCAGCAGAAAACAAACCCCAACAGGGAUUUAACUUGUUGCACAGU ..(((((......(((((..(((((.....))))).)))))...((((.....))))..)))))...(((((((.....(((.(((....))).)))...)))))))..... ( -23.40) >DroSec_CAF1 77538 109 + 1 AUAUAGCAAUAAAGAGCUCAGGCAAAUAUUUUGCUAAGCUUAUAUUUCCUUGCGAAAAAGUUAUCCAG---CAGAAAACAAACCCCAACAGGGAUUUAACUUGUUGCACAGU .....(((((((.(((((..(((((.....))))).))))).((..((((((.......(((.((...---..)).))).........))))))..))..)))))))..... ( -23.39) >DroSim_CAF1 78278 109 + 1 AUAUAGCAAUAAAGAGCUCAGGCAAAUAUUUUGCUAAGCUUAUAUUUCCUUGCGAAAAAGUUAUCCAG---CAGAAAACAAACCCCAACAGGGAUUUAACUUGUUGCACAGU .....(((((((.(((((..(((((.....))))).))))).((..((((((.......(((.((...---..)).))).........))))))..))..)))))))..... ( -23.39) >DroEre_CAF1 83291 109 + 1 AUAUAGCAAUAAAGAGCUCAGGCAAAUAUUUUGCUAAGCUUAUAUUUUCUUGCGAAAAAGUUAUCCAG---CAGAAAACAAACCCCAACAGGGAUUUAACUUGUUGCACAGC .....(((((((.(((((..(((((.....))))).)))))...((((((.((..............)---))))))).....(((....))).......)))))))..... ( -24.44) >DroYak_CAF1 83820 109 + 1 AUACAGCAAUAAAGAGCUCAGGCAAAUAUUUUGCUAAGCUUAUAUUUUCUUGCGAAAAAGUUAUCCAG---CAGAAAACAAACCCCAACAGGGAUUUAACUUGUUGCACAGU .....(((((((.(((((..(((((.....))))).)))))...((((((.((..............)---))))))).....(((....))).......)))))))..... ( -24.44) >consensus AUAUAGCAAUAAAGAGCUCAGGCAAAUAUUUUGCUAAGCUUAUAUUUCCUUGCGAAAAAGUUAUCCAG___CAGAAAACAAACCCCAACAGGGAUUUAACUUGUUGCACAGU .....(((((((.(((((..(((((.....))))).))))).((..((((((.......(((.((........)).))).........))))))..))..)))))))..... (-22.47 = -22.23 + -0.24)


| Location | 17,789,114 – 17,789,226 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
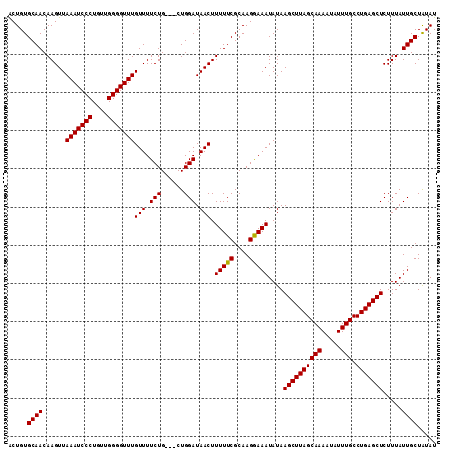
>3R_DroMel_CAF1 17789114 112 - 27905053 ACUGUGCAACAAGUUAAAUCCCUGUUGGGGUUUGUUUUCUGCUGCUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAU .....((((.....((((((((....)))))))).(((((..(((..((......))..))).)))))....((((((((((.....))).))))))).....))))..... ( -27.80) >DroSec_CAF1 77538 109 - 1 ACUGUGCAACAAGUUAAAUCCCUGUUGGGGUUUGUUUUCUG---CUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAU .....((((......(((((((....)))))))((((((((---(..((......))..)).)))))))...((((((((((.....))).))))))).....))))..... ( -27.10) >DroSim_CAF1 78278 109 - 1 ACUGUGCAACAAGUUAAAUCCCUGUUGGGGUUUGUUUUCUG---CUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAU .....((((......(((((((....)))))))((((((((---(..((......))..)).)))))))...((((((((((.....))).))))))).....))))..... ( -27.10) >DroEre_CAF1 83291 109 - 1 GCUGUGCAACAAGUUAAAUCCCUGUUGGGGUUUGUUUUCUG---CUGGAUAACUUUUUCGCAAGAAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAU ((...(((......((((((((....)))))))).....))---)..(((((..(((((....)))))....((((((((((.....))).)))))))..)))))))..... ( -27.80) >DroYak_CAF1 83820 109 - 1 ACUGUGCAACAAGUUAAAUCCCUGUUGGGGUUUGUUUUCUG---CUGGAUAACUUUUUCGCAAGAAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUGUAU ...(((((.(((...(((((((....)))))))((((((((---(..((......))..)).)))))))...((((((((((.....))).))))))).....))).))))) ( -28.90) >consensus ACUGUGCAACAAGUUAAAUCCCUGUUGGGGUUUGUUUUCUG___CUGGAUAACUUUUUCGCAAGGAAAUAUAAGCUUAGCAAAAUAUUUGCCUGAGCUCUUUAUUGCUAUAU .....((((......(((((((....)))))))(((.(((......))).))).(((((....)))))....((((((((((.....))).))))))).....))))..... (-27.34 = -27.10 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:05 2006