
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,781,534 – 17,781,651 |
| Length | 117 |
| Max. P | 0.976678 |

| Location | 17,781,534 – 17,781,625 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 73.82 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -15.54 |
| Energy contribution | -16.71 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17781534 91 + 27905053 GCUUU--UGCUAUUUCUGCUGCCAAAAUGCUCAACAAAACAUGUCGAAAAUGUGCCACAUGCCAGUAGCAGGAGCAGCAGGAGCUGUAGGAGC------ (((((--(((..(((((((((((....((((..((....(((((.(........).)))))...)))))).).))))))))))..))))))))------ ( -32.20) >DroGri_CAF1 65865 86 + 1 GGUUU--UGUUGCUGUUGUUGCCAAAUUGCAC-ACAAAACAUGUCGAAAAUGUGCCAUAUGCCAGCAGCAAAUGAAAUC---GU-UGUCUGUU------ (((((--..(((((((((..((...((.((((-(................))))).))..))))))))).))..)))))---..-........------ ( -21.39) >DroSec_CAF1 70180 91 + 1 GCUUU--UGCUAUUUCUGCUGCCAAAAUGCUCAACAAAACAUGUCGAAAAUGUGCCACAUGCCAGAAGCAGGAGCAGCACGAGCUGUAGGAGC------ (((((--(((..(((.(((((((....((((........(((((.(........).))))).....)))).).)))))).)))..))))))))------ ( -26.22) >DroEre_CAF1 75722 82 + 1 GCUUU--UGCUAUUUCUGCUGCCAAAAUGCUCAACAAAACAUGUCGAAAAUGUGCCACAUGCCAGUAGCAGGAGCAGCAGGAGC--------------- (((((--((((.((((((((((........((.(((.....))).))...((.((.....)))))))))))))).)))))))))--------------- ( -28.30) >DroYak_CAF1 75729 99 + 1 GCUUUUCUGCUAUUUCUGCUGCCAAAAUGCUCAACAAAACAUGUCGAAAAUGUGCCACAUGCCAGUAGCAGGAGCAGCAGGAGCUGUAGGAGCUGUUGU ((((..((((..(((((((((((....((((..((....(((((.(........).)))))...)))))).).))))))))))..))))))))...... ( -31.10) >DroMoj_CAF1 81984 77 + 1 -----------GUUAUUGUUGCCAAAAUGCAC-ACAAAACAUGUCGAAAAUGUGCCAUGUGCCAGCAGCAAAUGGAAUC---GU-UGUCUGCU------ -----------(((.(((((((......))).-)))))))..((.((.((((..((((.(((.....))).))))...)---))-).)).)).------ ( -17.30) >consensus GCUUU__UGCUAUUUCUGCUGCCAAAAUGCUCAACAAAACAUGUCGAAAAUGUGCCACAUGCCAGUAGCAGGAGCAGCAGGAGC_GUAGGAGC______ ((((...((((.((((((((((......((........((((.......)))).......))..)))))))))).)))).))))............... (-15.54 = -16.71 + 1.17)



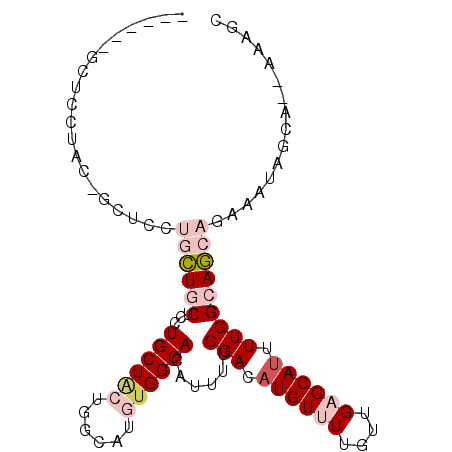
| Location | 17,781,534 – 17,781,625 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 73.82 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -13.81 |
| Energy contribution | -15.20 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17781534 91 - 27905053 ------GCUCCUACAGCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCAGCAGAAAUAGCA--AAAGC ------.........(((.((((((((..((((((......))))))....(((((((.....)))))))......))))))))....))).--..... ( -29.10) >DroGri_CAF1 65865 86 - 1 ------AACAGACA-AC---GAUUUCAUUUGCUGCUGGCAUAUGGCACAUUUUCGACAUGUUUUGU-GUGCAAUUUGGCAACAACAGCAACA--AAACC ------........-..---.......((((.((((((((((..(.((((.......)))).)..)-))))...(((....)))))))).))--))... ( -22.80) >DroSec_CAF1 70180 91 - 1 ------GCUCCUACAGCUCGUGCUGCUCCUGCUUCUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCAGCAGAAAUAGCA--AAAGC ------(((((....((((((((..(...(((.....))).)..))))......((((.....)))))))).....)).)))..........--..... ( -26.60) >DroEre_CAF1 75722 82 - 1 ---------------GCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCAGCAGAAAUAGCA--AAAGC ---------------(((.((((((((..((((((......))))))....(((((((.....)))))))......))))))))....))).--..... ( -28.90) >DroYak_CAF1 75729 99 - 1 ACAACAGCUCCUACAGCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCAGCAGAAAUAGCAGAAAAGC .....((((.....)))).((((((((..((((((......))))))....(((((((.....)))))))......))))))))............... ( -29.10) >DroMoj_CAF1 81984 77 - 1 ------AGCAGACA-AC---GAUUCCAUUUGCUGCUGGCACAUGGCACAUUUUCGACAUGUUUUGU-GUGCAUUUUGGCAACAAUAAC----------- ------(((((.((-(.---........)))))))).(((((..(.((((.......)))).)..)-))))...(((....)))....----------- ( -20.80) >consensus ______GCUCCUAC_GCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCAGCAGAAAUAGCA__AAAGC ....................((((((...((((((......))))))......(((.((((((....)))))).)))))))))................ (-13.81 = -15.20 + 1.39)


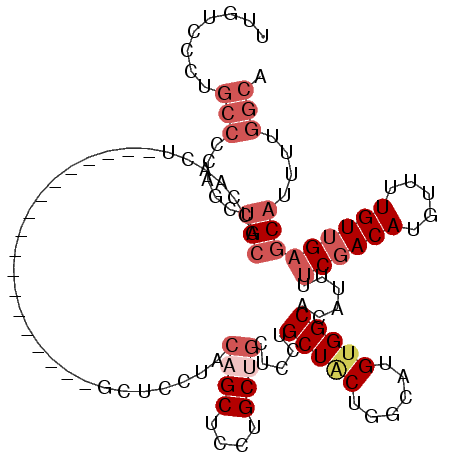
| Location | 17,781,551 – 17,781,651 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -13.28 |
| Energy contribution | -15.46 |
| Covariance contribution | 2.18 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17781551 100 - 27905053 UUGUCCCUGCCCCAGCUGCACCAACU------------------GCUCCUACAGCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCA ........(((...((..(((((...------------------((.....((((....)))).....))...)))..))..)).....(((((((.....)))))))......))). ( -27.80) >DroSec_CAF1 70197 100 - 1 UUGUCCCUGCCCCAGCUGCACCUACU------------------GCUCCUACAGCUCGUGCUGCUCCUGCUUCUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCA ........(((..((..(((.....)------------------))..))...((((((((..(...(((.....))).)..))))......((((.....)))))))).....))). ( -28.60) >DroEre_CAF1 75739 91 - 1 UUGUCCCUGCCCCAGCUGCCCCAGCU---------------------------GCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCA ........(((.((((((...)))))---------------------------).......(((((.((((((......)))))).......((((.....)))))))))....))). ( -26.90) >DroWil_CAF1 90382 75 - 1 -----------------GCAGCAACC------------------GGUAGAACA-----U--GGCUCUGGCUGCUGGCAUGUGGCACAUUUUCGACAUGUUUUGU-GCACAUUUUGGCA -----------------.(((((.((------------------((.((....-----.--..)))))).)))))((..((((((((..............)))-)).)))....)). ( -23.24) >DroYak_CAF1 75748 118 - 1 UUGUCCCUGCCCCAGCUGCCCCAACUUCCCCAGCUGCCACAACAGCUCCUACAGCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCA ........(((..(((((............)))))((((((..........((((....))))....(((.....))))))))).....(((((((.....)))))))......))). ( -31.70) >consensus UUGUCCCUGCCCCAGCUGCACCAACU__________________GCUCCUACAGCUCCUGCUGCUCCUGCUACUGGCAUGUGGCACAUUUUCGACAUGUUUUGUUGAGCAUUUUGGCA ........(((.....(((................................((((....)))).....(((((......)))))......((((((.....)))))))))....))). (-13.28 = -15.46 + 2.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:00 2006