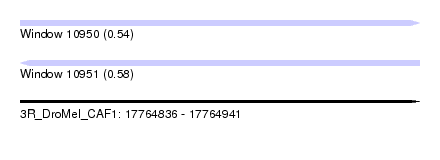
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,764,836 – 17,764,941 |
| Length | 105 |
| Max. P | 0.576881 |

| Location | 17,764,836 – 17,764,941 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -16.58 |
| Consensus MFE | -13.08 |
| Energy contribution | -13.41 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
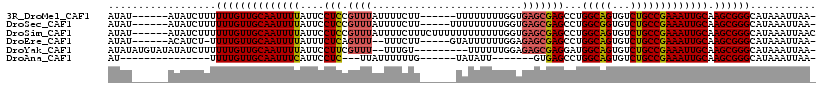
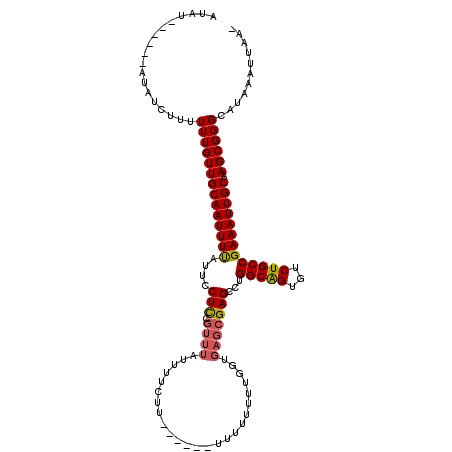
>3R_DroMel_CAF1 17764836 105 + 27905053 -UUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACUGCCAGGCUCGCUCACCAAAAAAAA------AAGAAAAUAAACGGAGGAAUAAAAUUGCAACAAAAAAGAUAU------AUAU -.............(.(((((((((.(((((...)))))....((.(((...........------.............)))))...))))))))))...........------.... ( -15.96) >DroSec_CAF1 53345 106 + 1 -UUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACCGCCAGGCUCGCUCACCAAAAAAAAA-----AAGAAAAUAAACGGAGGAAUAAAAUUGCAACAAAAAAGAUAU------AUAU -.............(.(((((((((.(((.......)))....((.(((............-----.............)))))...))))))))))...........------.... ( -13.01) >DroSim_CAF1 54209 112 + 1 GUUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACUGCCAGGCUCGCUCACCAAAAAAAAAAAAGAAAGAAAAUAAACGGAGGAAUAAAAUUGCAACAAAAAAGAUAU------AUAU ((...(((((.((((.(((...(((((((((...))))).((........))............)))).....))).))).)..)))))...))..............------.... ( -15.90) >DroEre_CAF1 56006 103 + 1 -UUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACUGCCAGGCUCGCUCUCCAAAAAAUAC-----AAGAAA--AAACUGAGAAAUAAAAUUGCAACAAAA-AGAUGU------AUAU -......(((((...((((((((((.(((((...)))))........((((..........-----......--.....))))....))))))))......-))..))------))). ( -16.40) >DroYak_CAF1 57364 106 + 1 -UUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACUGCCAUCCUCGCUCUCCAAAAAA---------ACAAA--AAACGAAGGAAUAAAAUUGCAACAAAAAAGAUAUAUACAUAUAU -.............(.(((((((((.(((((...))))).((((((............---------.....--...)).))))...))))))))))..................... ( -18.45) >DroAna_CAF1 61792 86 + 1 -UUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACUGCCAGGCUCAC-------AAUAUA------CAAAAAAUAA---GAGGAAUGAAAUUGCAACAAAA---------------AU -.............(.(((((((((((((((...)))))...(((..-------......------..........---)))....)))))))))))....---------------.. ( -19.77) >consensus _UUAAUUUAUGCCCGCUUGCAAUUUCGGCAGACACUGCCAGGCUCGCUCACCAAAAAAAA______AAGAAAAUAAACGGAGGAAUAAAAUUGCAACAAAAAAGAUAU______AUAU ..............(.(((((((((.(((((...)))))...(((..................................))).....))))))))))..................... (-13.08 = -13.41 + 0.33)

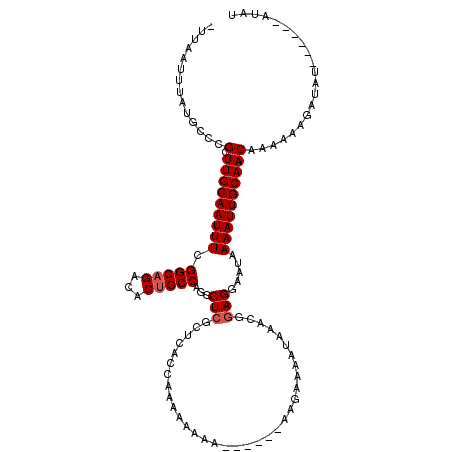
| Location | 17,764,836 – 17,764,941 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.31 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17764836 105 - 27905053 AUAU------AUAUCUUUUUUGUUGCAAUUUUAUUCCUCCGUUUAUUUUCUU------UUUUUUUUGGUGAGCGAGCCUGGCAGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAA- ....------........((((((((((((((.......((((((((.....------........)))))))).....(((((...))))))))))))).))))))..........- ( -22.52) >DroSec_CAF1 53345 106 - 1 AUAU------AUAUCUUUUUUGUUGCAAUUUUAUUCCUCCGUUUAUUUUCUU-----UUUUUUUUUGGUGAGCGAGCCUGGCGGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAA- ....------........((((((((((((((.......((((((((.....-----.........)))))))).....(((((...))))))))))))).))))))..........- ( -21.74) >DroSim_CAF1 54209 112 - 1 AUAU------AUAUCUUUUUUGUUGCAAUUUUAUUCCUCCGUUUAUUUUCUUUCUUUUUUUUUUUUGGUGAGCGAGCCUGGCAGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAAC ....------........((((((((((((((.......((((((((...................)))))))).....(((((...))))))))))))).))))))........... ( -22.11) >DroEre_CAF1 56006 103 - 1 AUAU------ACAUCU-UUUUGUUGCAAUUUUAUUUCUCAGUUU--UUUCUU-----GUAUUUUUUGGAGAGCGAGCCUGGCAGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAA- ....------......-.((((((((((((((........((((--.(((((-----..........))))).))))..(((((...))))))))))))).))))))..........- ( -21.30) >DroYak_CAF1 57364 106 - 1 AUAUAUGUAUAUAUCUUUUUUGUUGCAAUUUUAUUCCUUCGUUU--UUUGU---------UUUUUUGGAGAGCGAGGAUGGCAGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAA- ...(((((...........(((((((((((((..((((.(((((--(..(.---------.....)..)))))))))).(((((...))))))))))))).))))))))))......- ( -29.70) >DroAna_CAF1 61792 86 - 1 AU---------------UUUUGUUGCAAUUUCAUUCCUC---UUAUUUUUUG------UAUAUU-------GUGAGCCUGGCAGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAA- ..---------------.((((((((((((((......(---((((..(...------...)..-------)))))...(((((...))))))))))))).))))))..........- ( -20.90) >consensus AUAU______AUAUCUUUUUUGUUGCAAUUUUAUUCCUCCGUUUAUUUUCUU______UUUUUUUUGGUGAGCGAGCCUGGCAGUGUCUGCCGAAAUUGCAAGCGGGCAUAAAUUAA_ ..................((((((((((((((....(((.((((.........................)))))))...(((((...))))))))))))).))))))........... (-17.06 = -17.31 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:51 2006